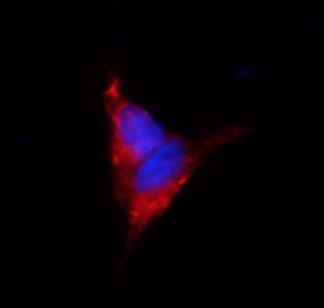
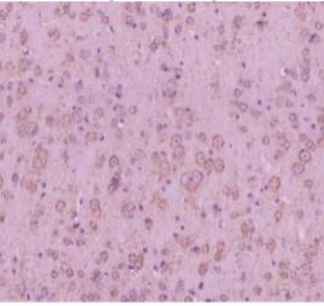
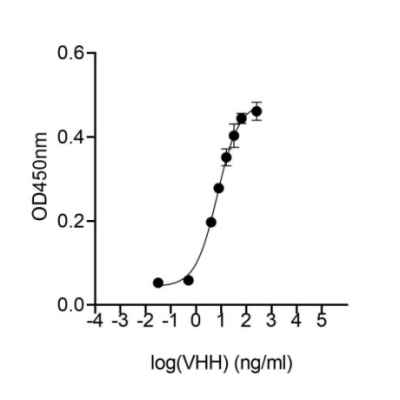
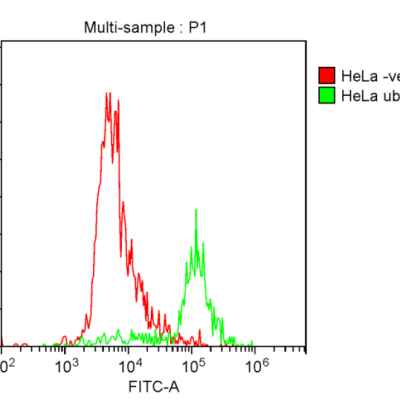
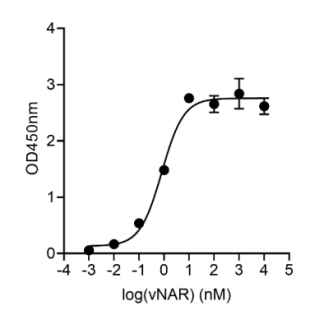
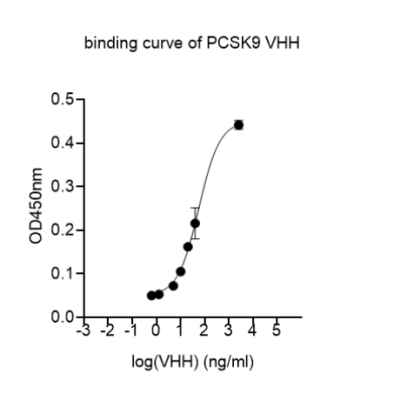
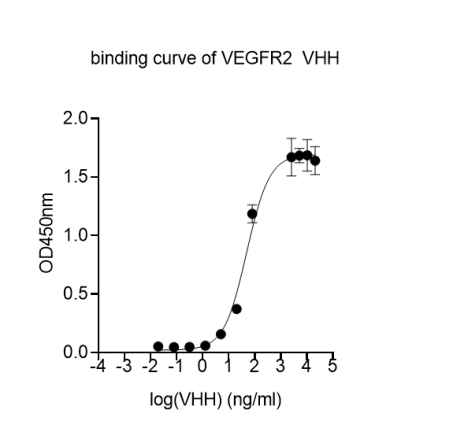
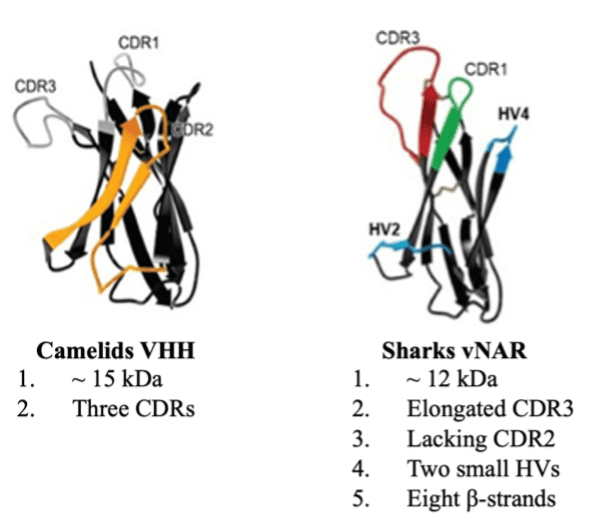
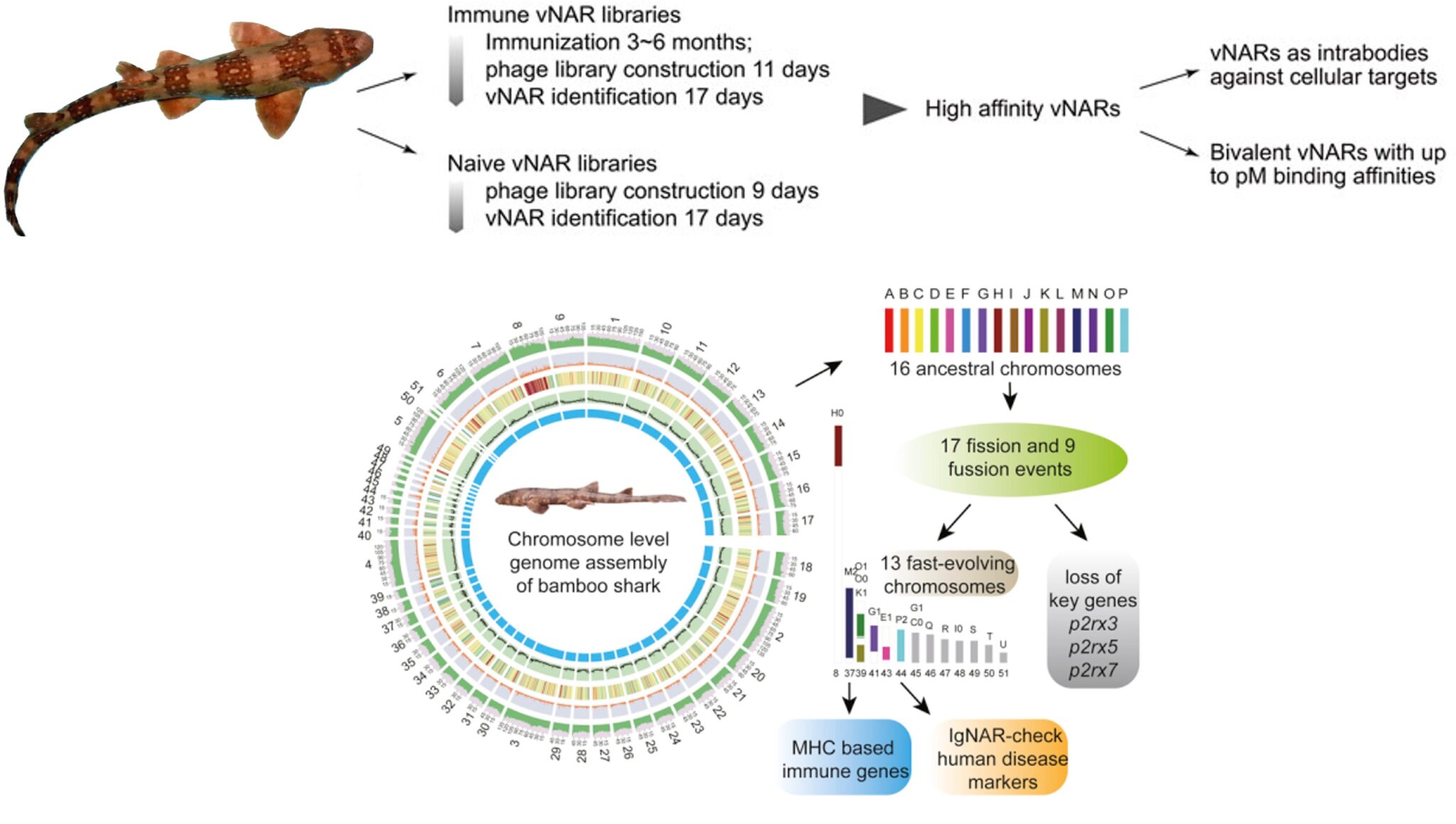
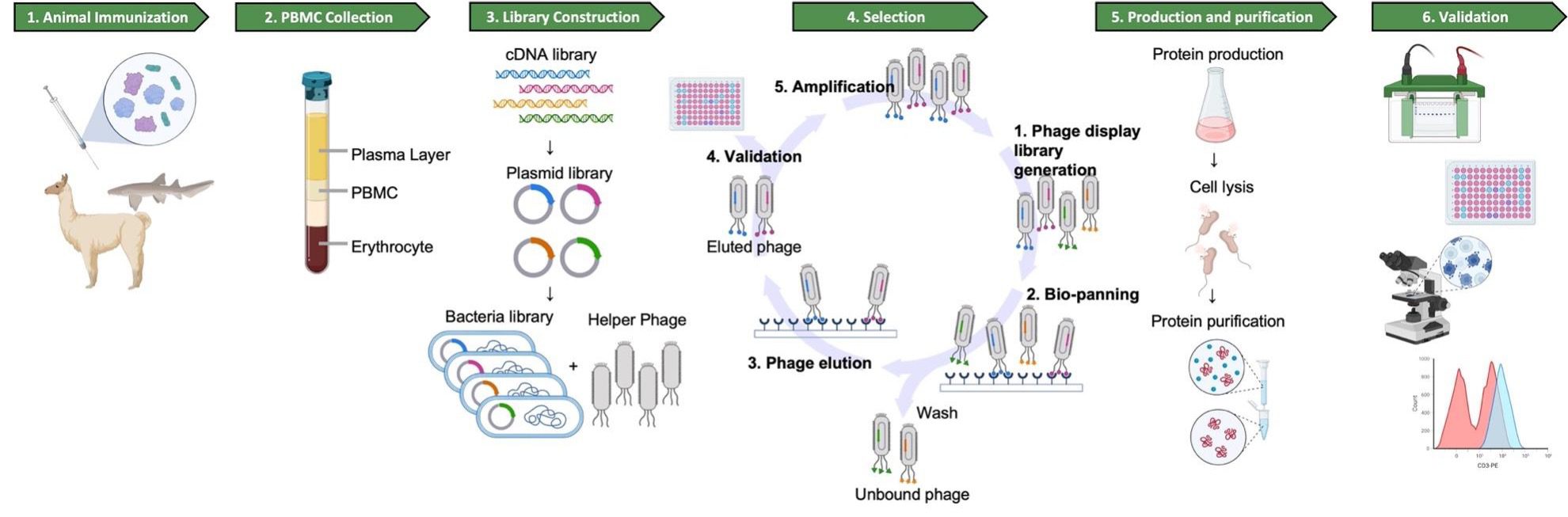
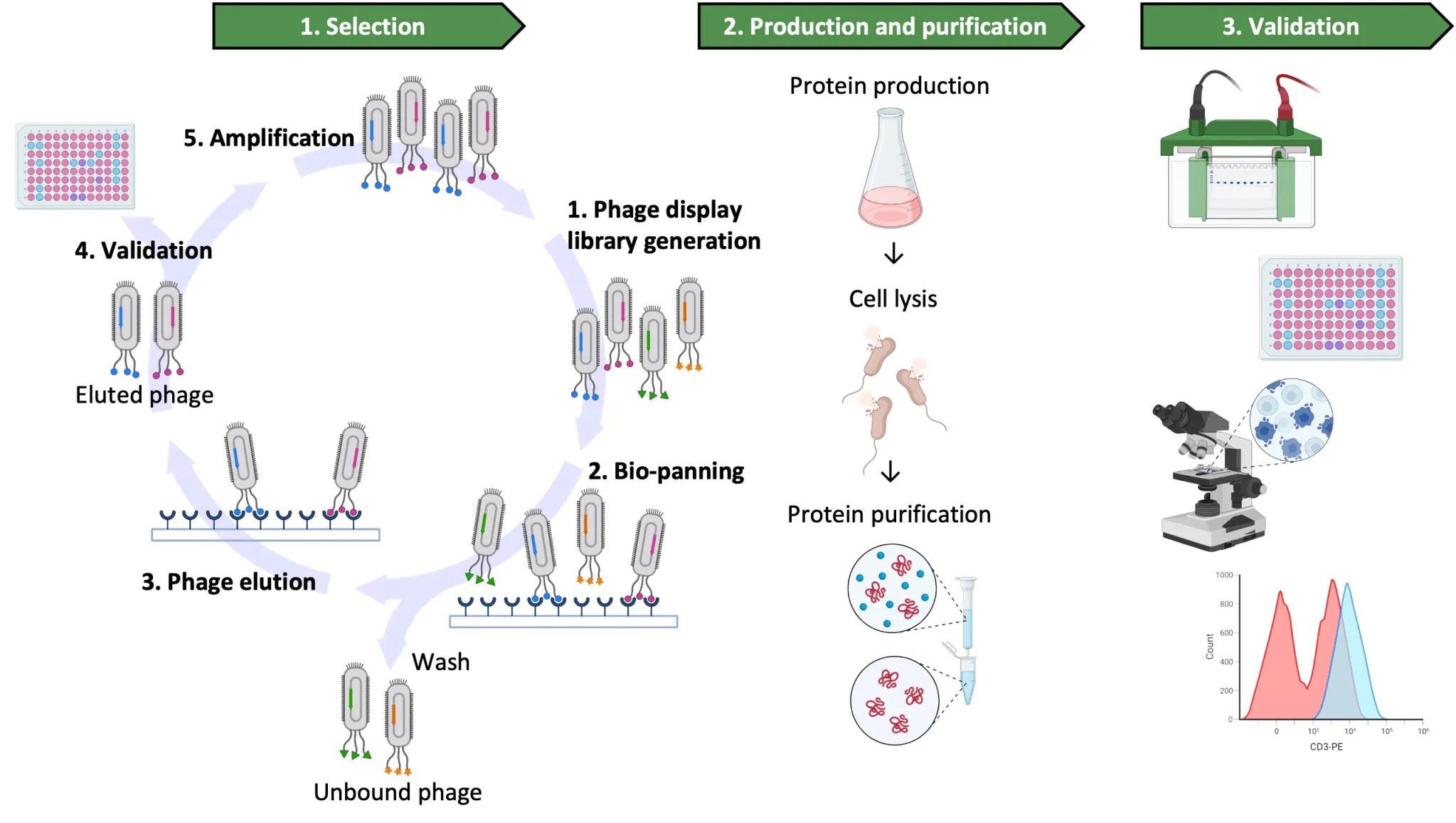
Jotbody (HK) Limited, established in 2020, is a biotech company specializing in nanobodies and extracellular vesicle (EV) research. We are driven by a team of passionate scientists and experienced board members. Located in Hong Kong and Shenzhen, we have access to thriving biotech ecosystems and state-of-the-art facilities.
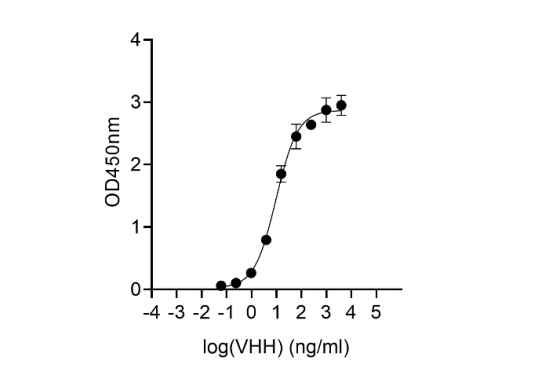
Our vision is to make high-quality research and development widely accessible, benefiting patients and healthcare providers globally. We take pride in our comprehensive nanobody CRO services, which support scientific research. Additionally, we offer top-notch, timely, and cost-effective nanobody-based research reagents to the global scientific community.
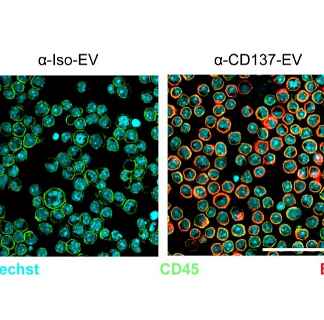
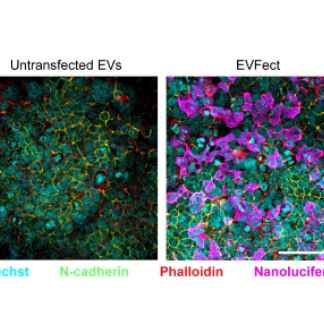
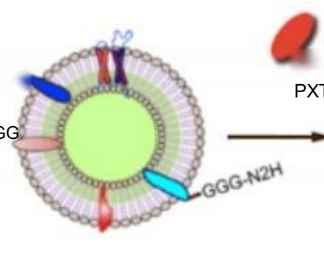
Moreover, our expertise extends beyond nanobodies as we possess specialized knowledge in the field of EV therapeutics. As pioneers in this field, we use advanced technologies and platforms to drive EV research and practical applications. Our expertise includes enhancing EVs with nanobodies, enabling precise and targeted therapeutic delivery. We provide a range of EV solutions, such as loading, surface modification, and validation for effective targeting.
At Jotbody, we are dedicated to advancing nanobodies and EV research, delivering innovative solutions for targeted therapeutic delivery, and contributing to scientific progress.