Histone ‘reader’ proteins contain evolutionarily conserved domains that bind to specific histone post-translational modifications (PTMs)1. These proteins have been shown to regulate diverse cellular processes via recruitment of epigenetic complexes or chromatin modifying / remodeling enzymes. Histone readers are an untapped wealth of information that underlie the foundation of chromatin regulation and could be used to advance therapeutics, diagnostics, and even personalized medicine. However, very little is known about how this diverse class of interactors functions in the context of chromatin.
Histone peptides: an incomplete chromatin model?
For years, histone peptide-based assays have been the gold standard for developing antibodies against PTMs, as well as defining the targets of chromatin modifying enzymes and histone readers. Scientists have depended on this data to build essential theories of chromatin function, as it relates to gene expression, DNA damage repair, and the development of human disease.
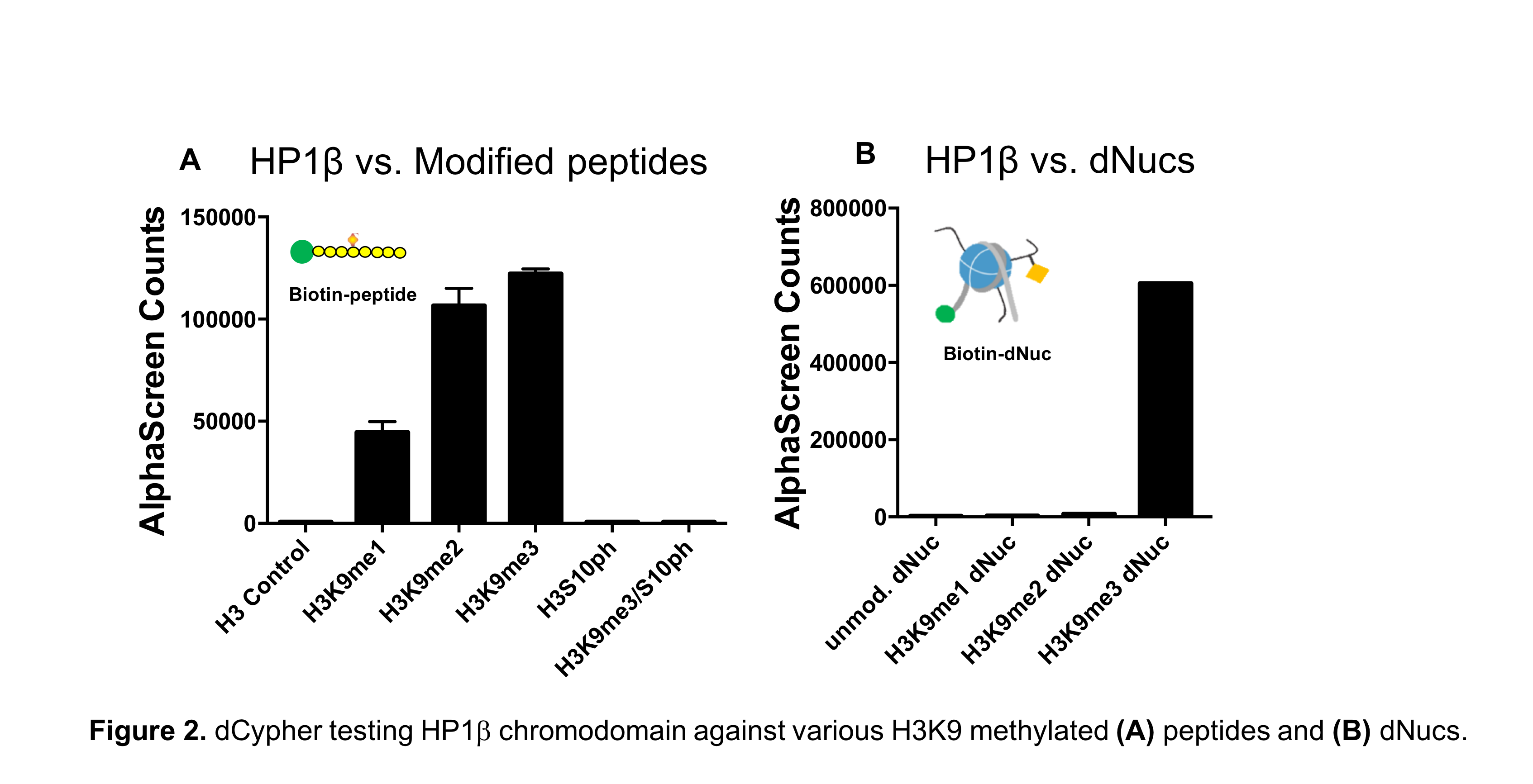
It has largely been assumed that reader interactions with histone PTMs occur independently of the larger nucleosomal structure, supporting the use of peptides or peptide arrays in chromatin studies. However, recent NMR spectroscopy data reveal that histone PTMs display substantial interactions with nucleosomal DNA, and that these contacts influence protein binding activity2-5. Furthermore, EpiCypher’s own SNAP-ChIP™ (Sample Normalization and Antibody Profiling for ChIP) platform has shown that histone peptides fail to accurately predict antibody binding specificity in ChIP experiments6. To learn more, see our recent Molecular Cell publication6.
All of this points to a misguided dependence on peptide-based data (such as histone peptide arrays), but direct comparison of peptides and nucleosomal substrates in chromatin reader assays has been difficult to achieve.
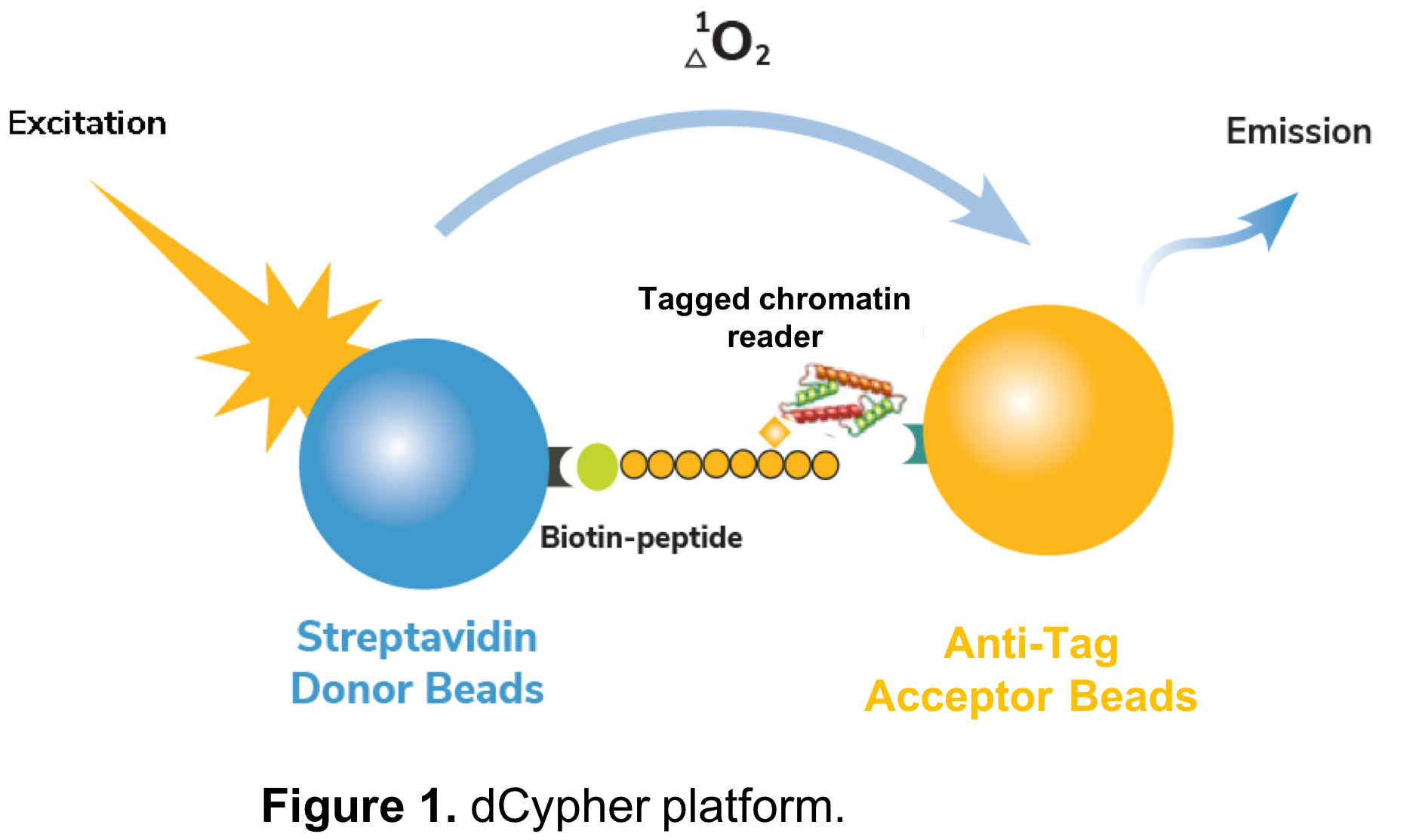
dCypher as an innovative platform to test histone peptides vs. nucleosomes