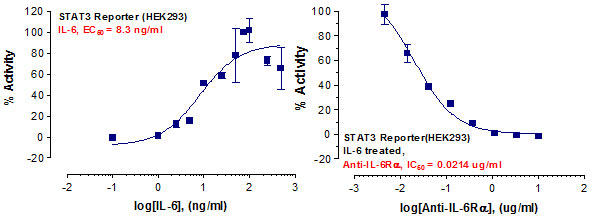
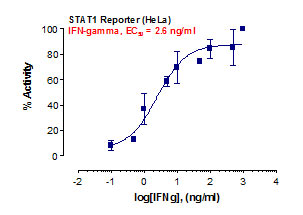
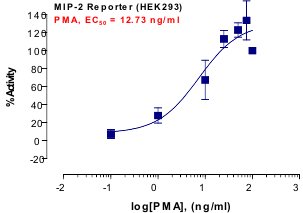
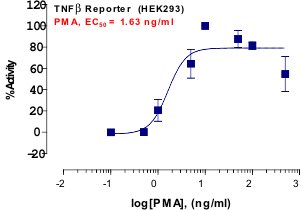
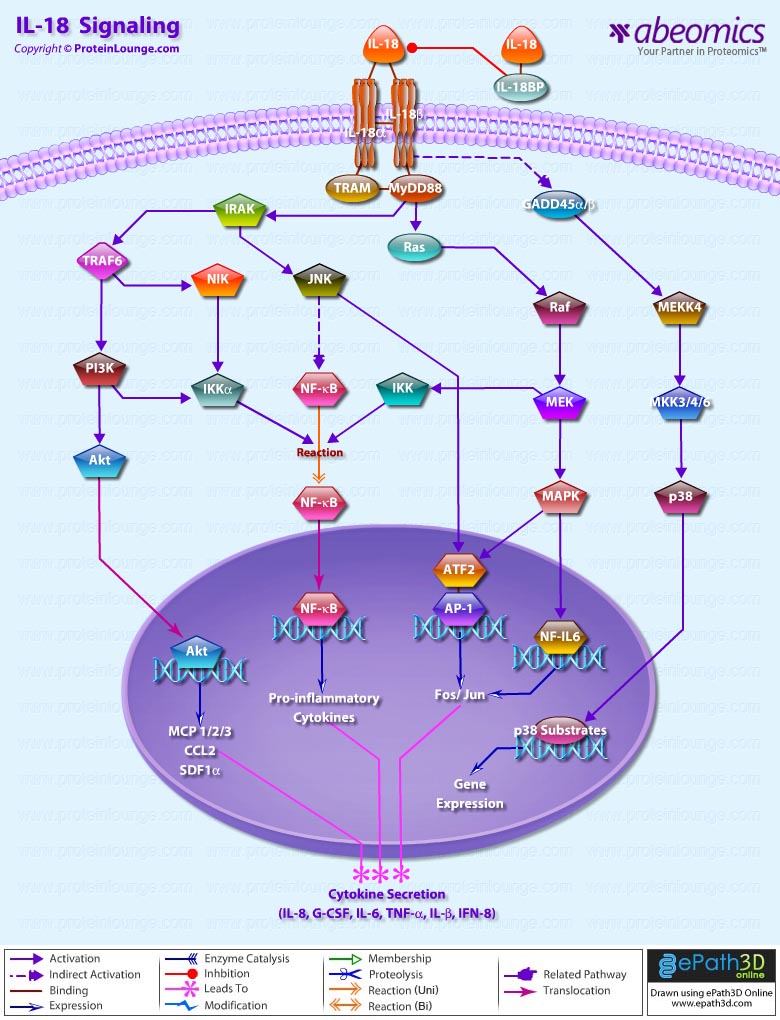
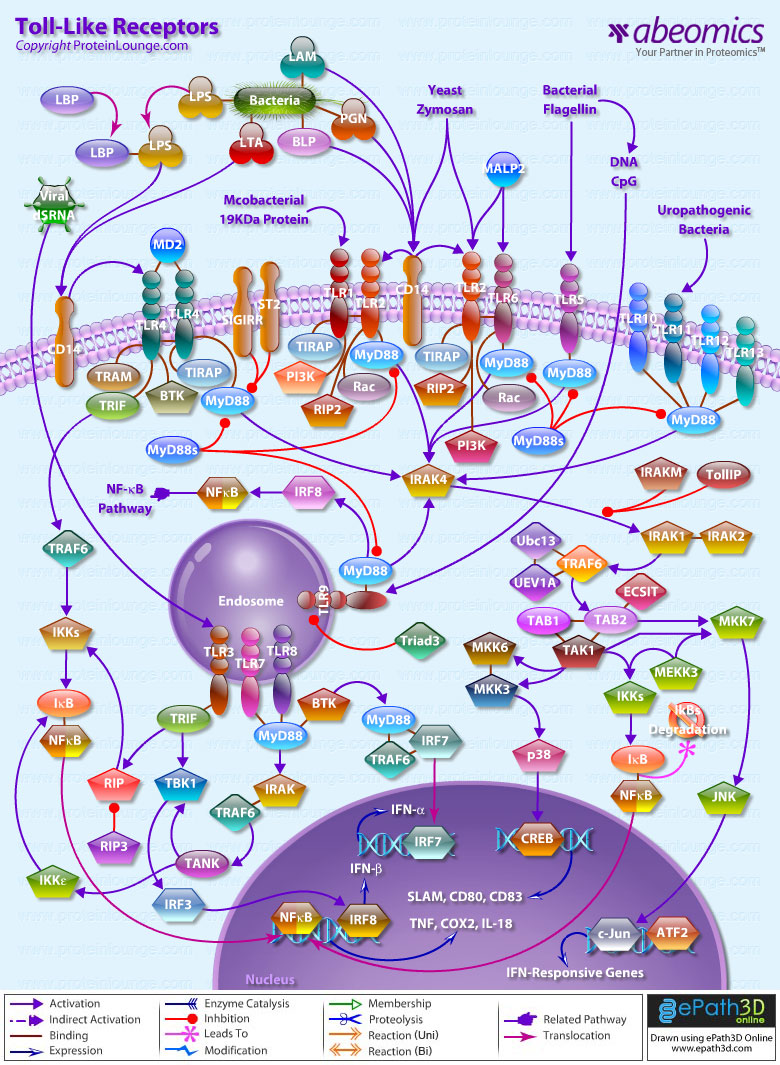
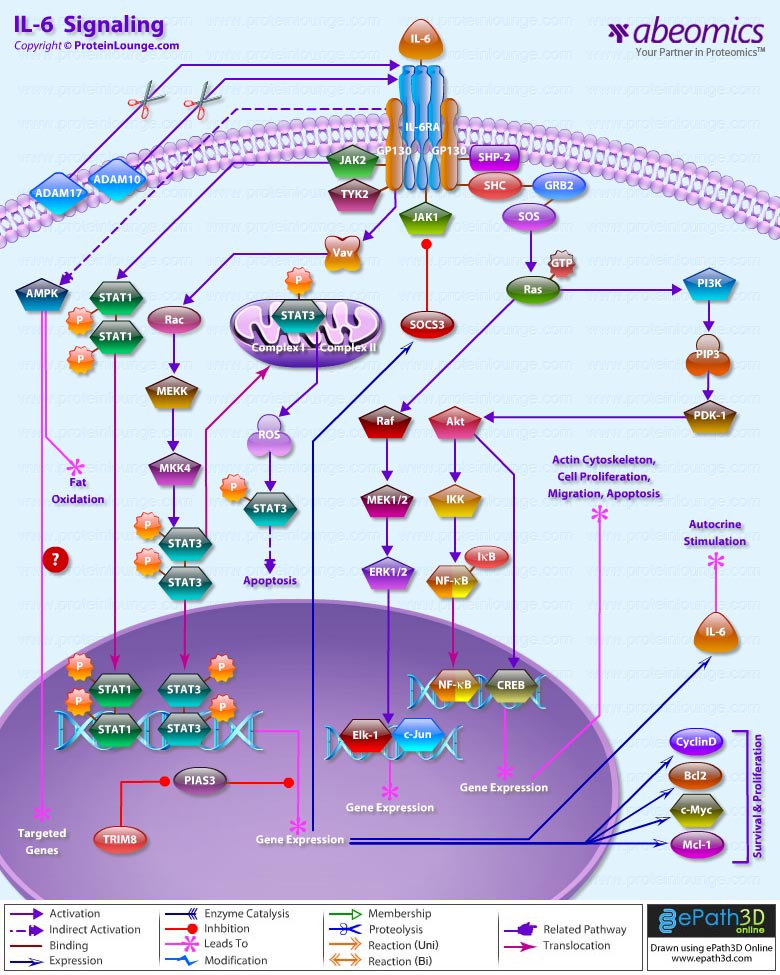
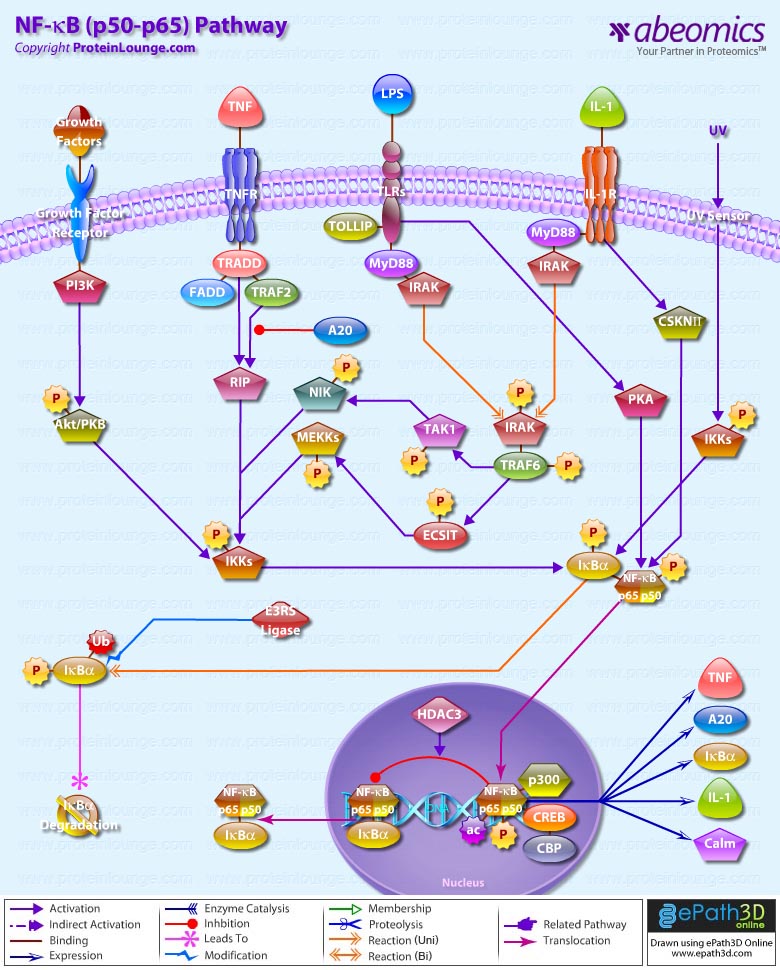
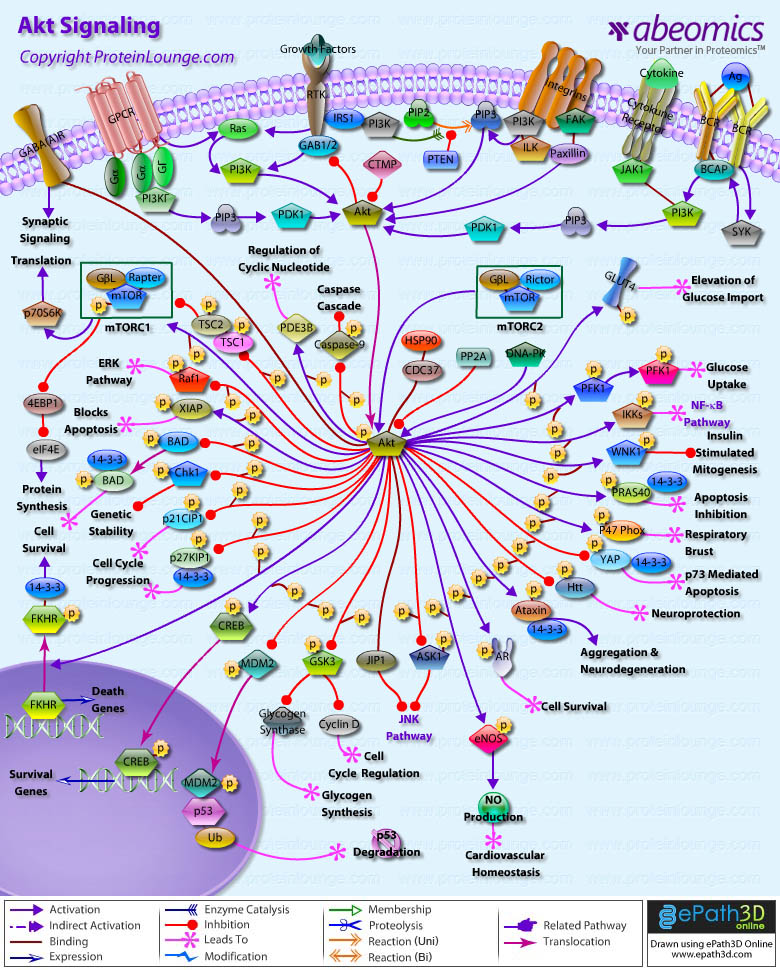
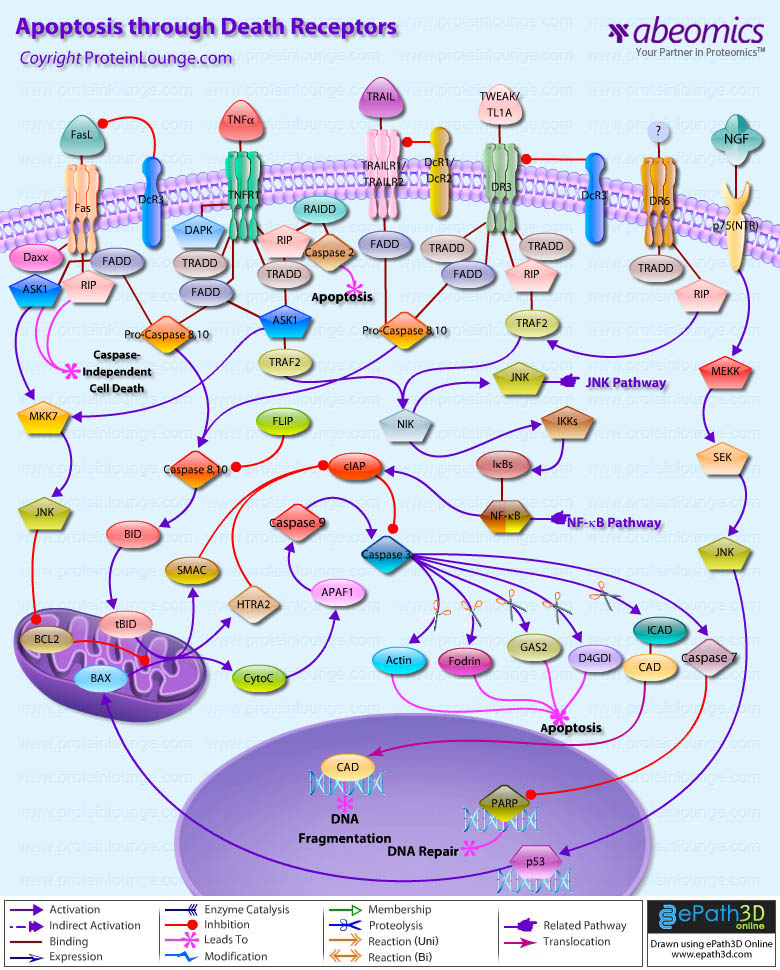
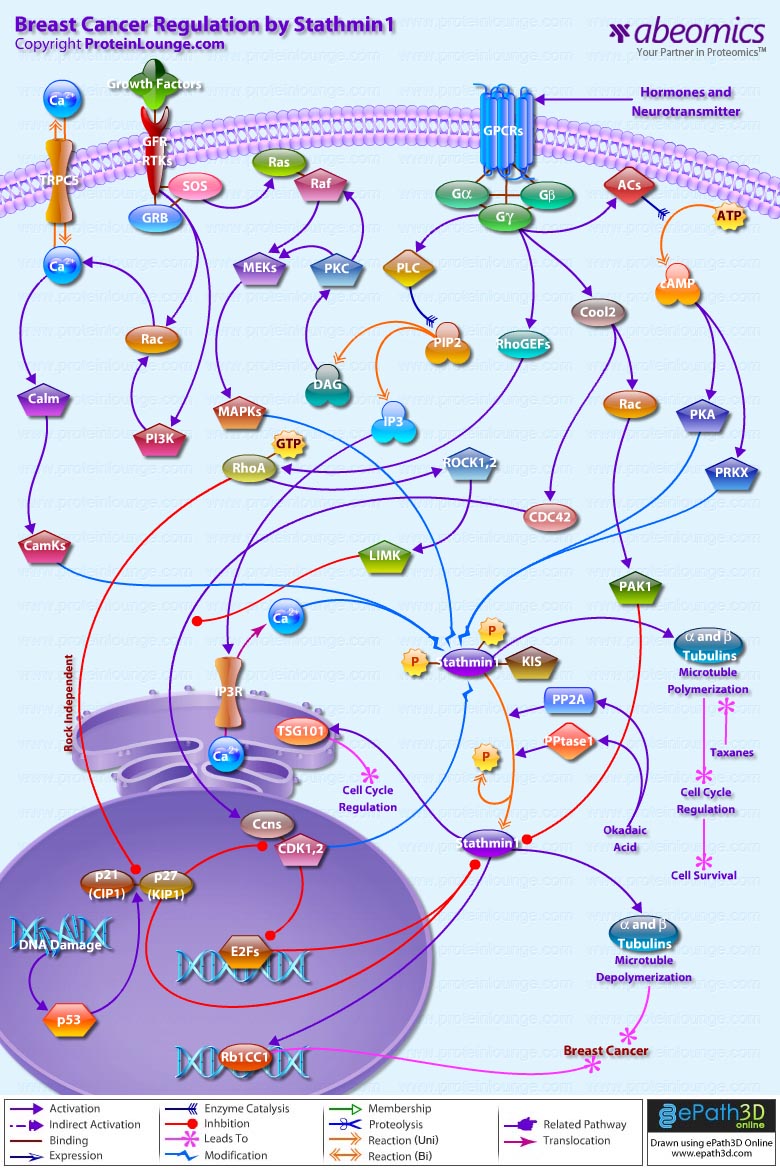
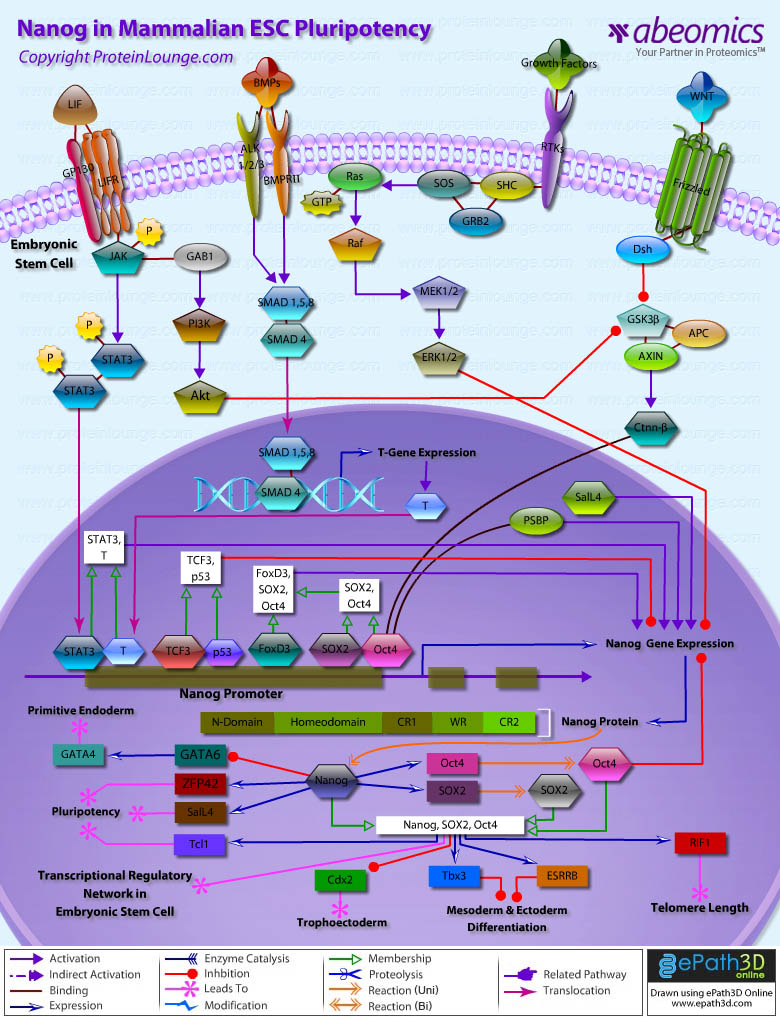
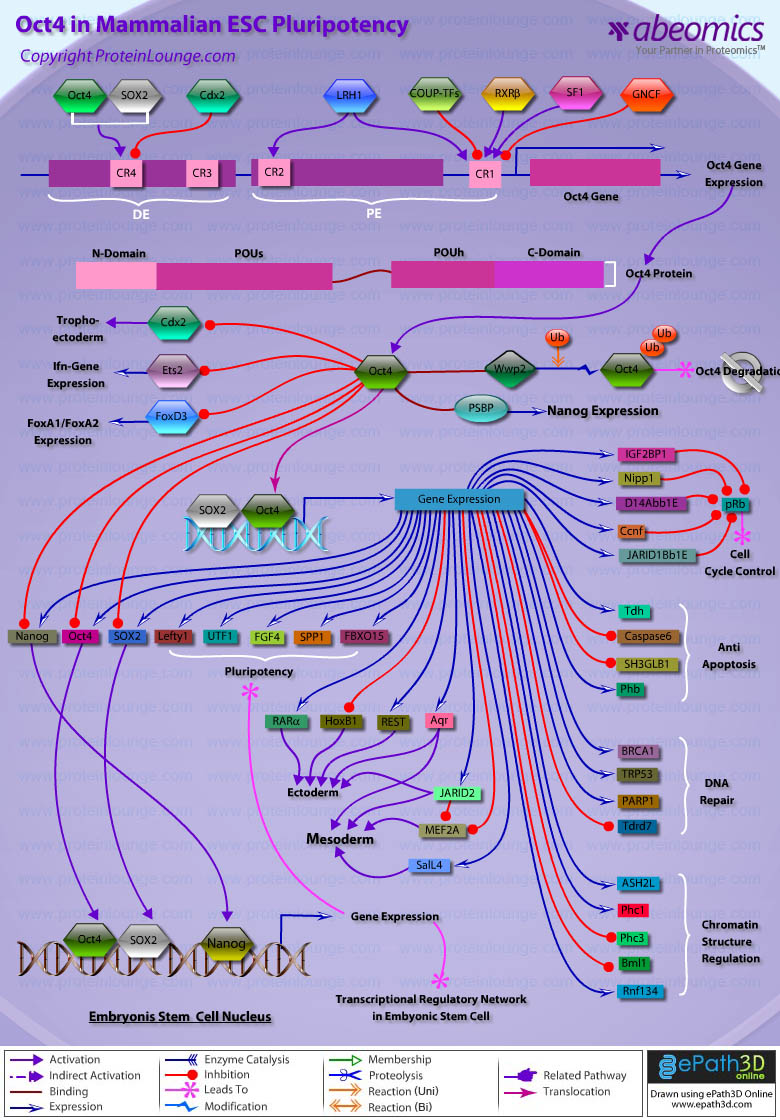
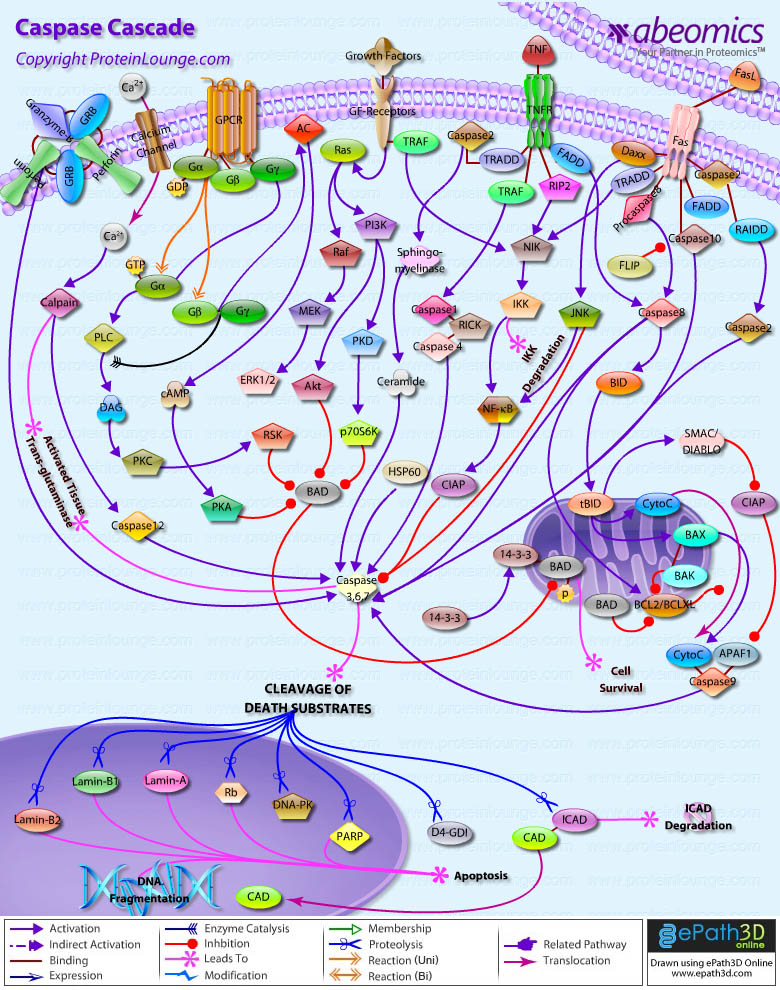
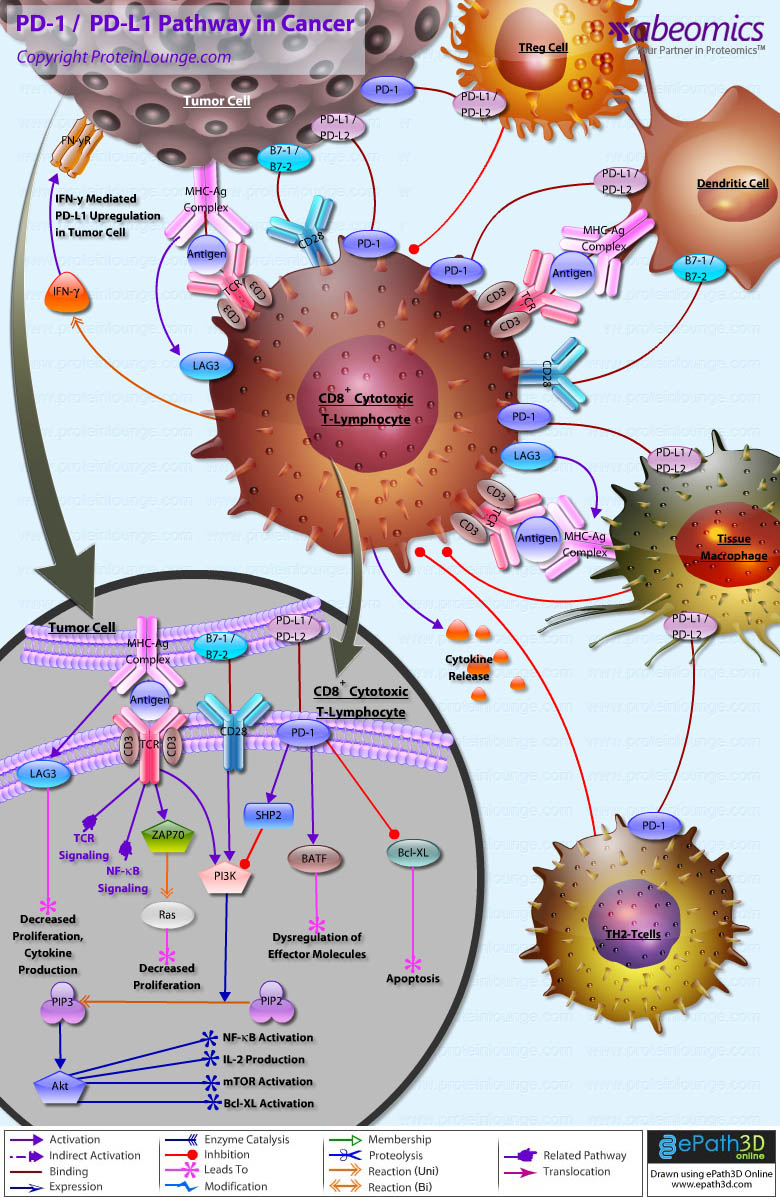
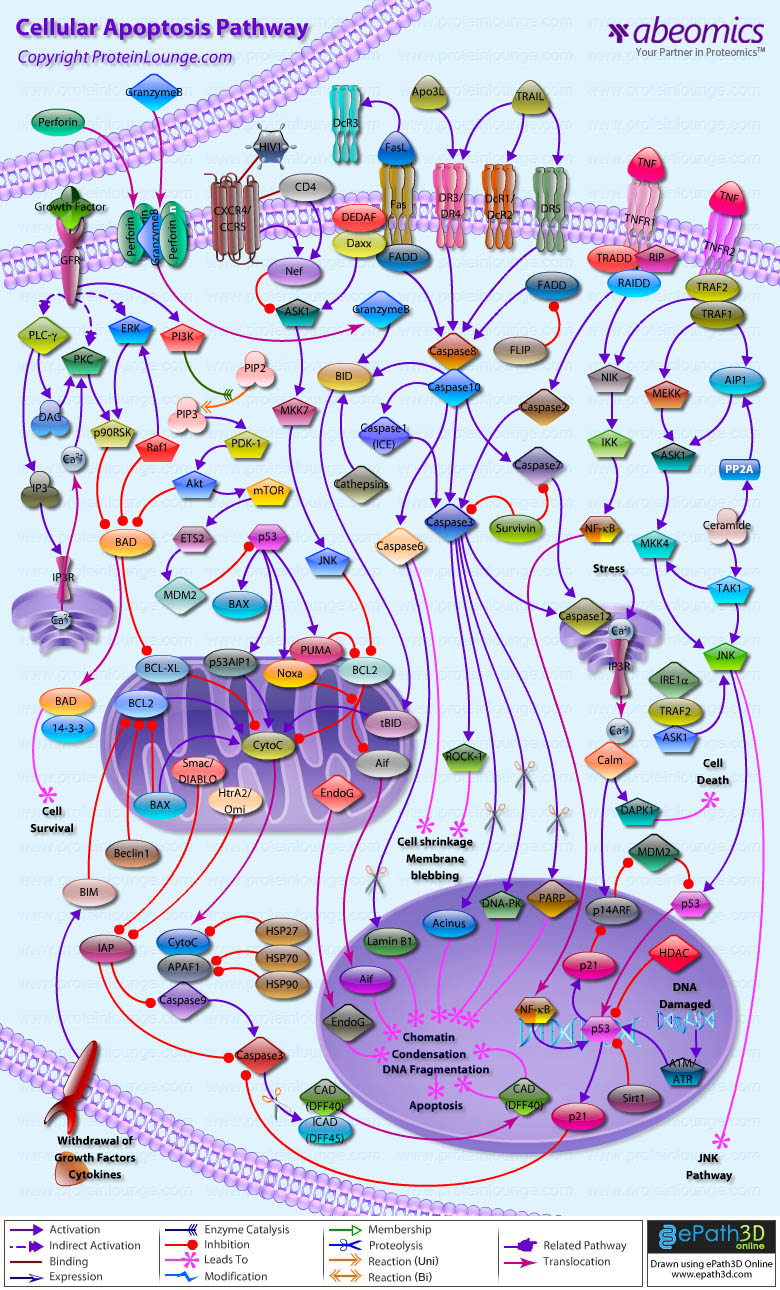
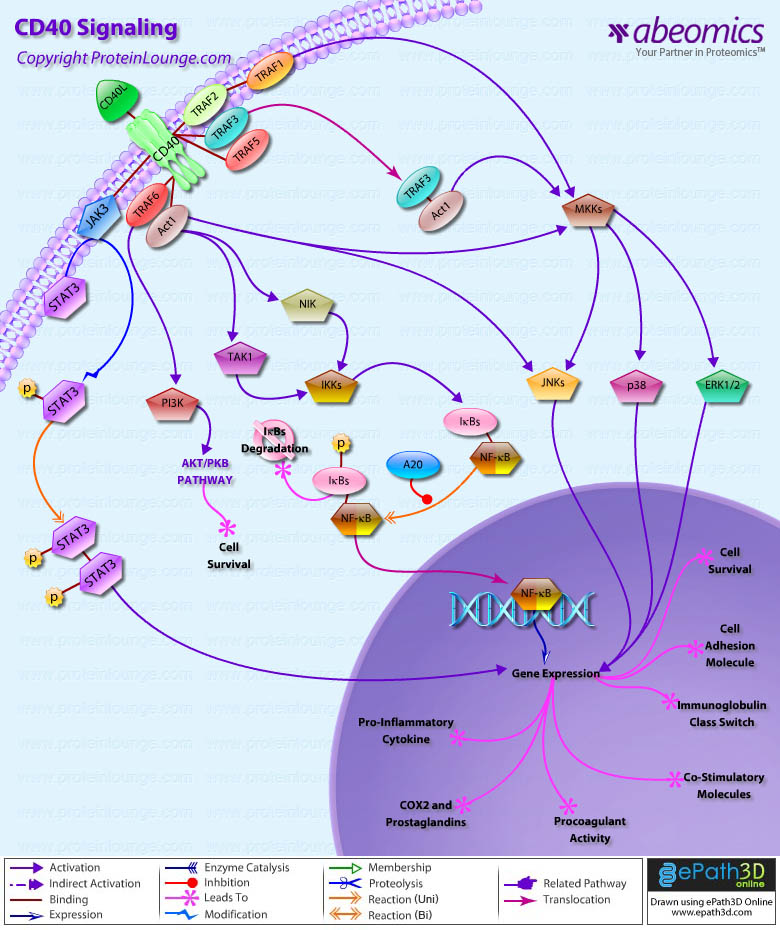
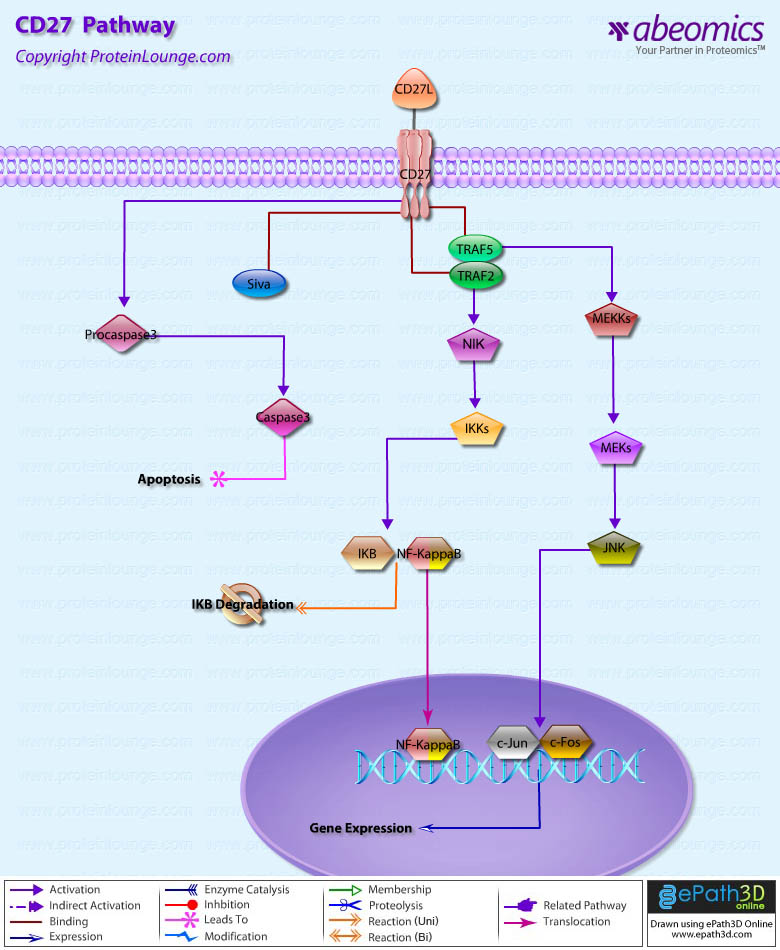
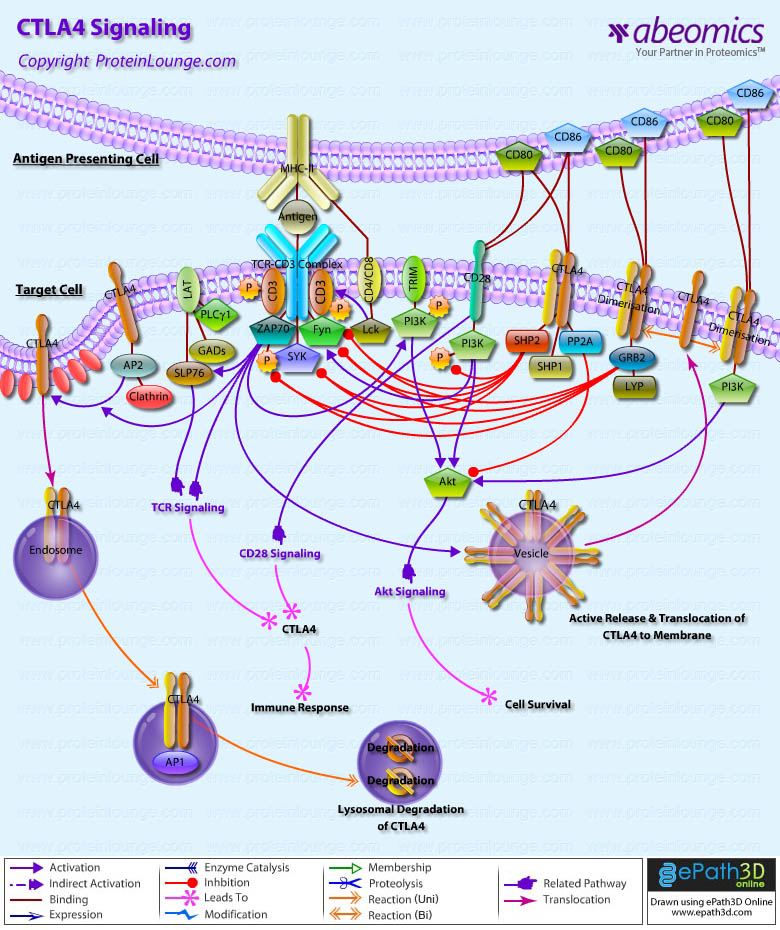
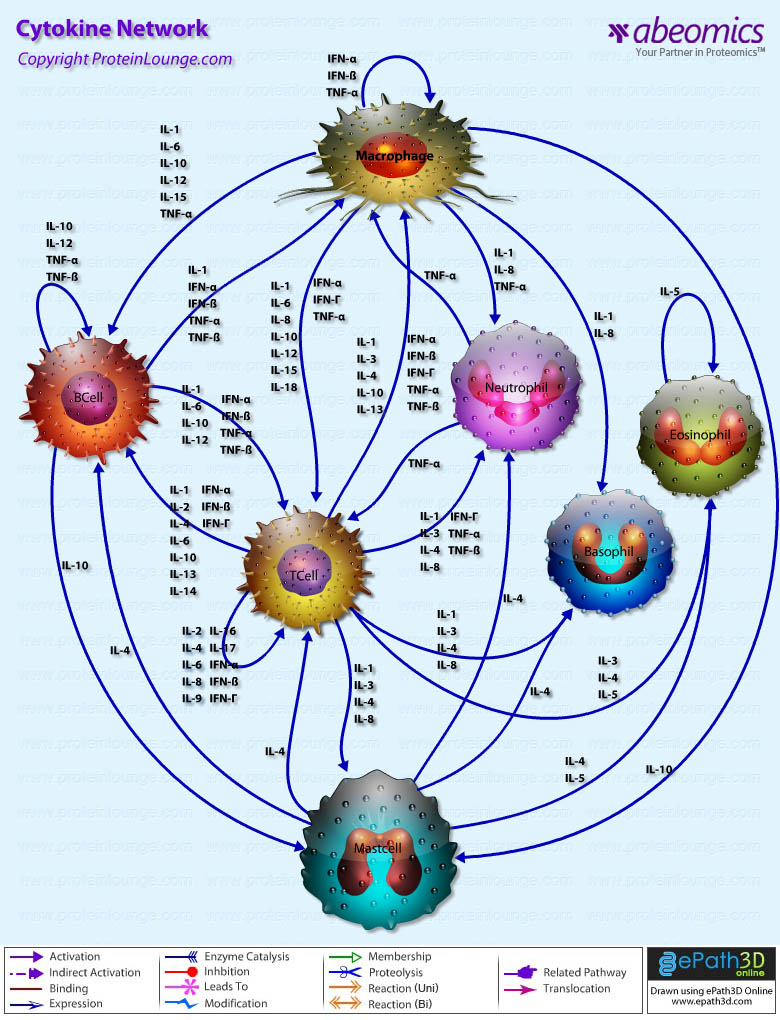
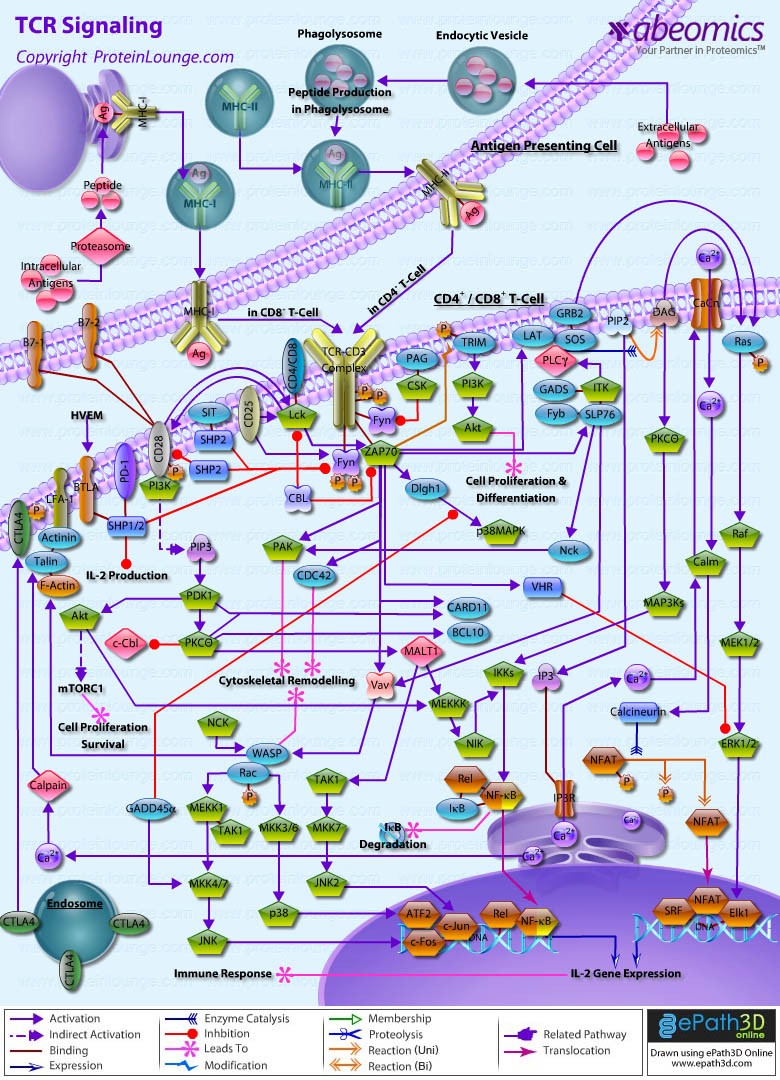
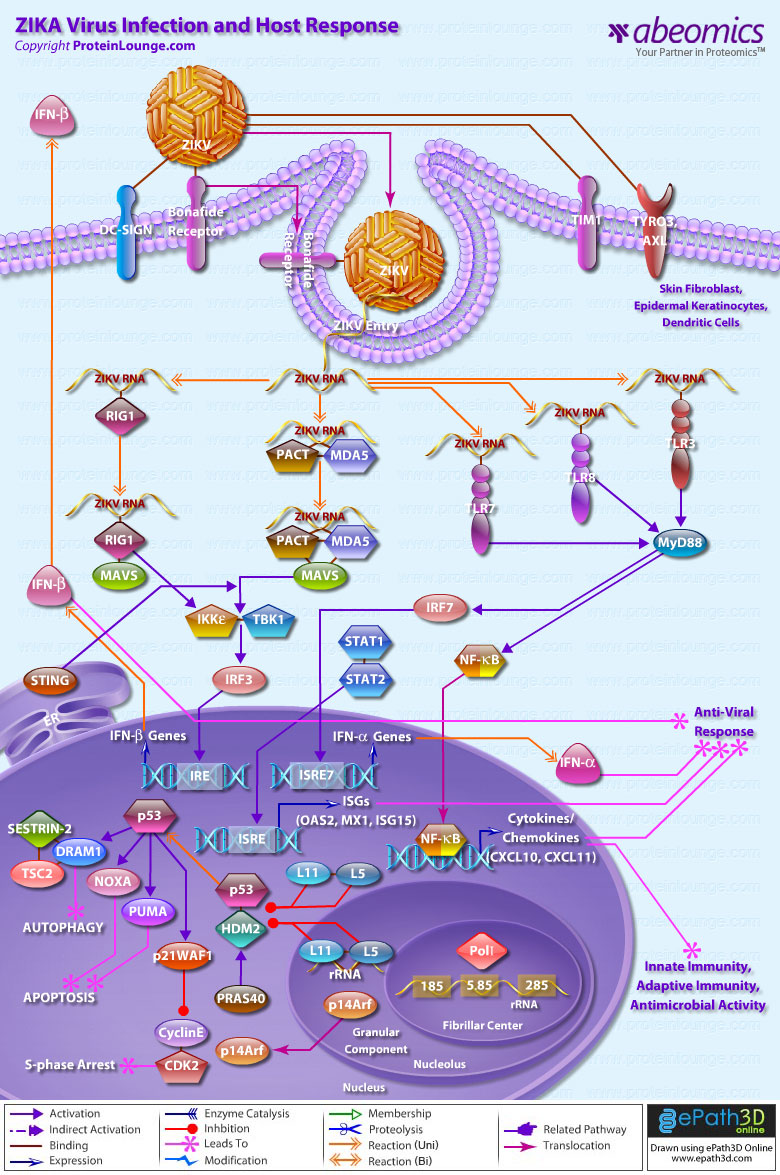
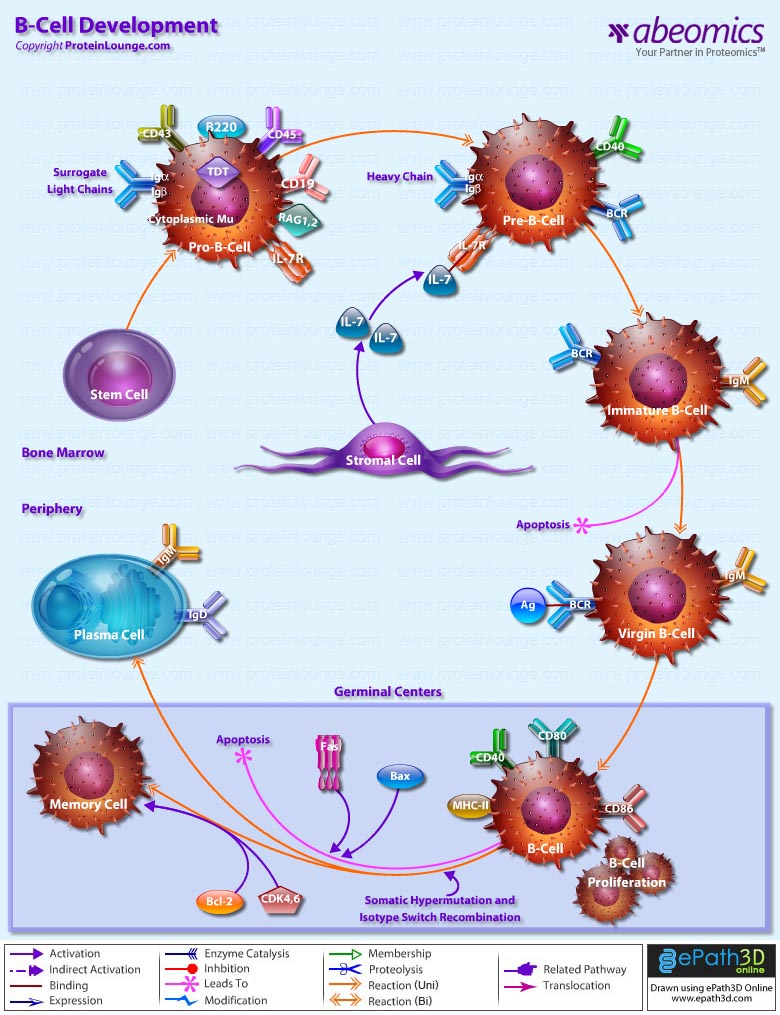
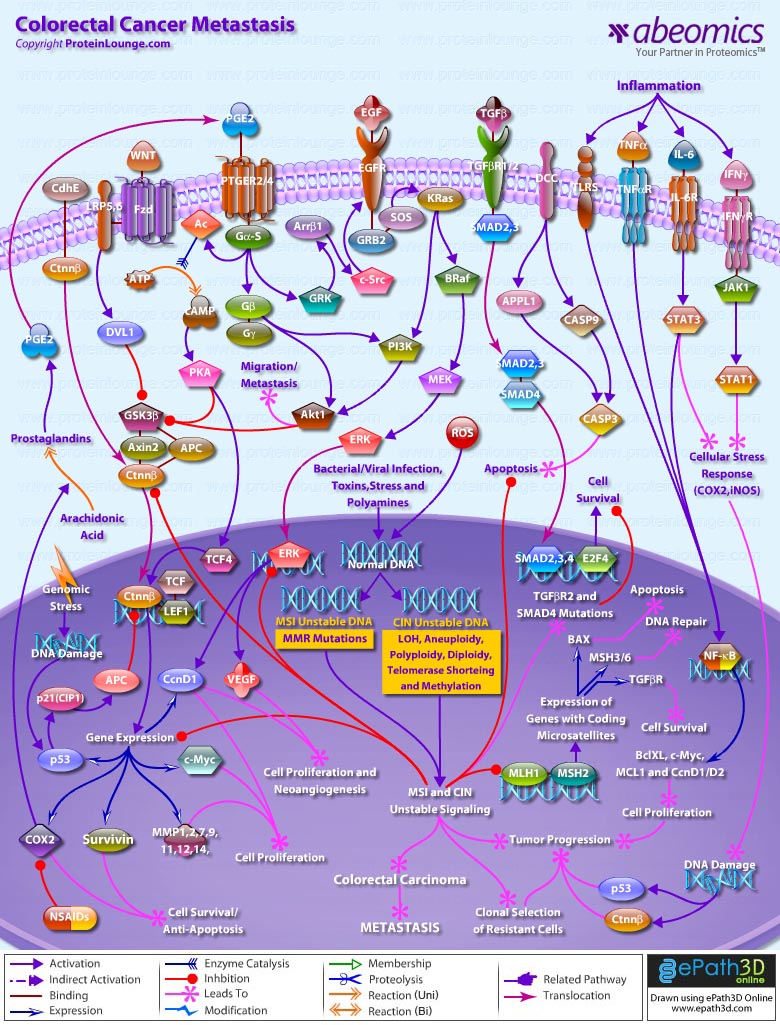
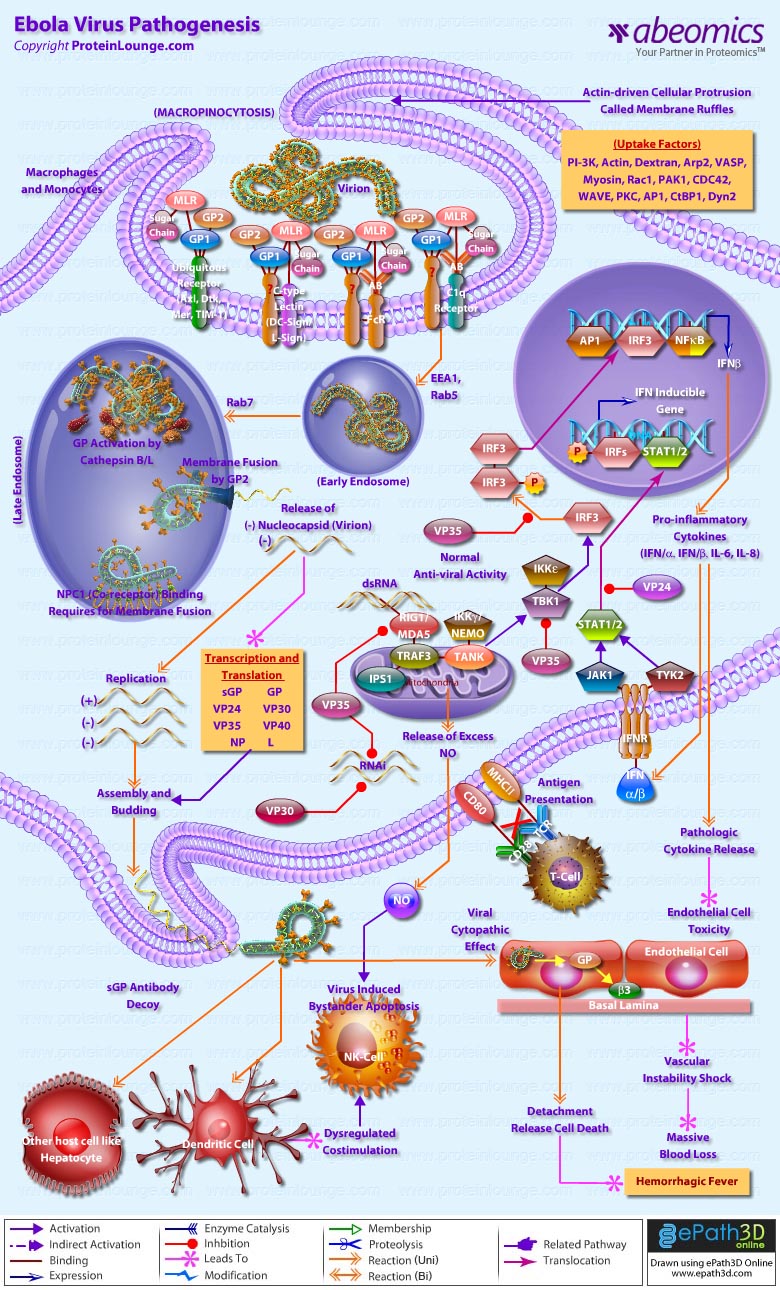
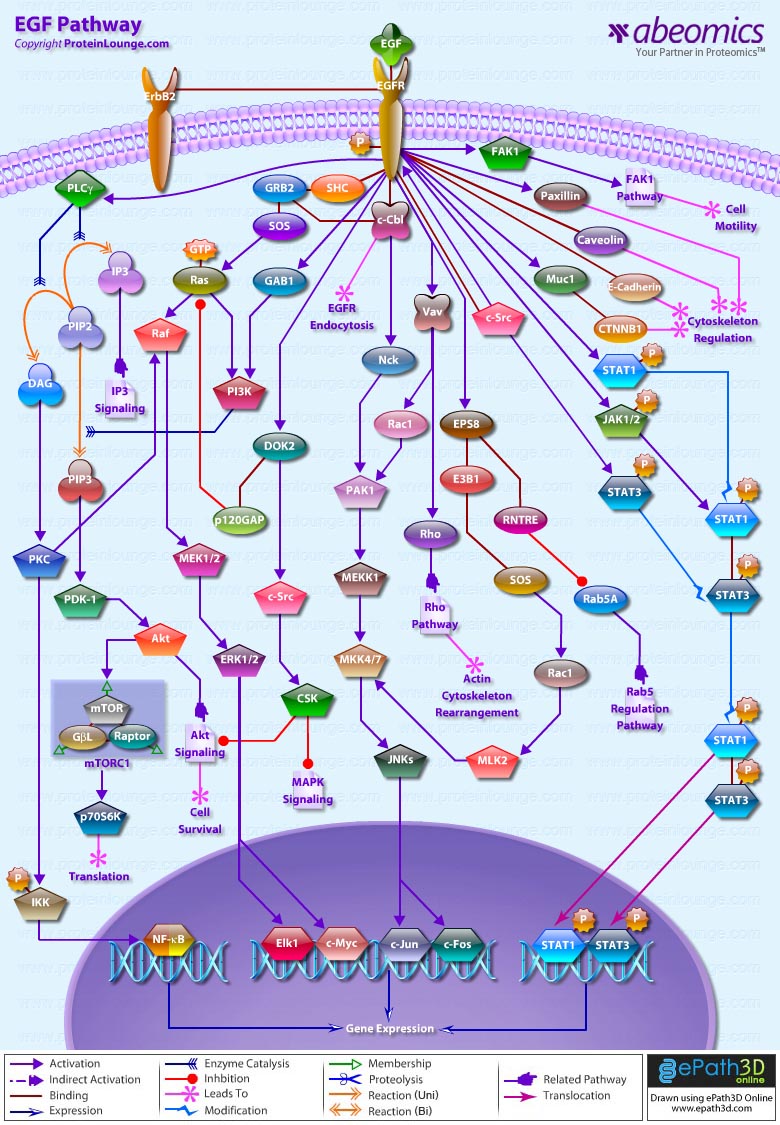
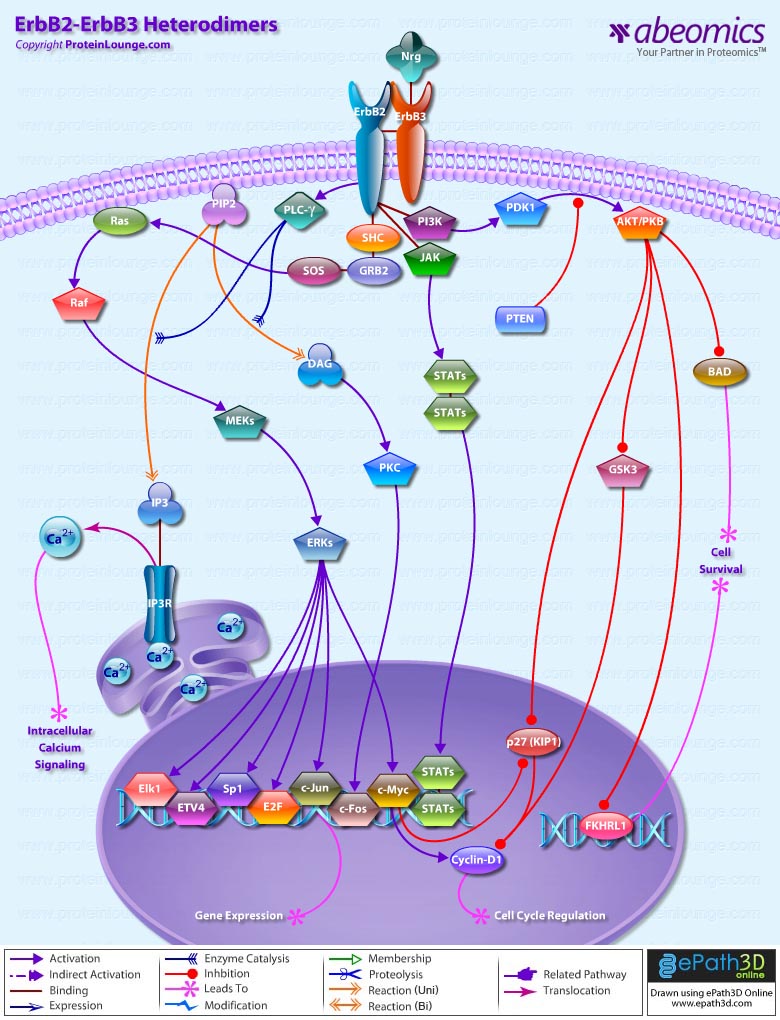
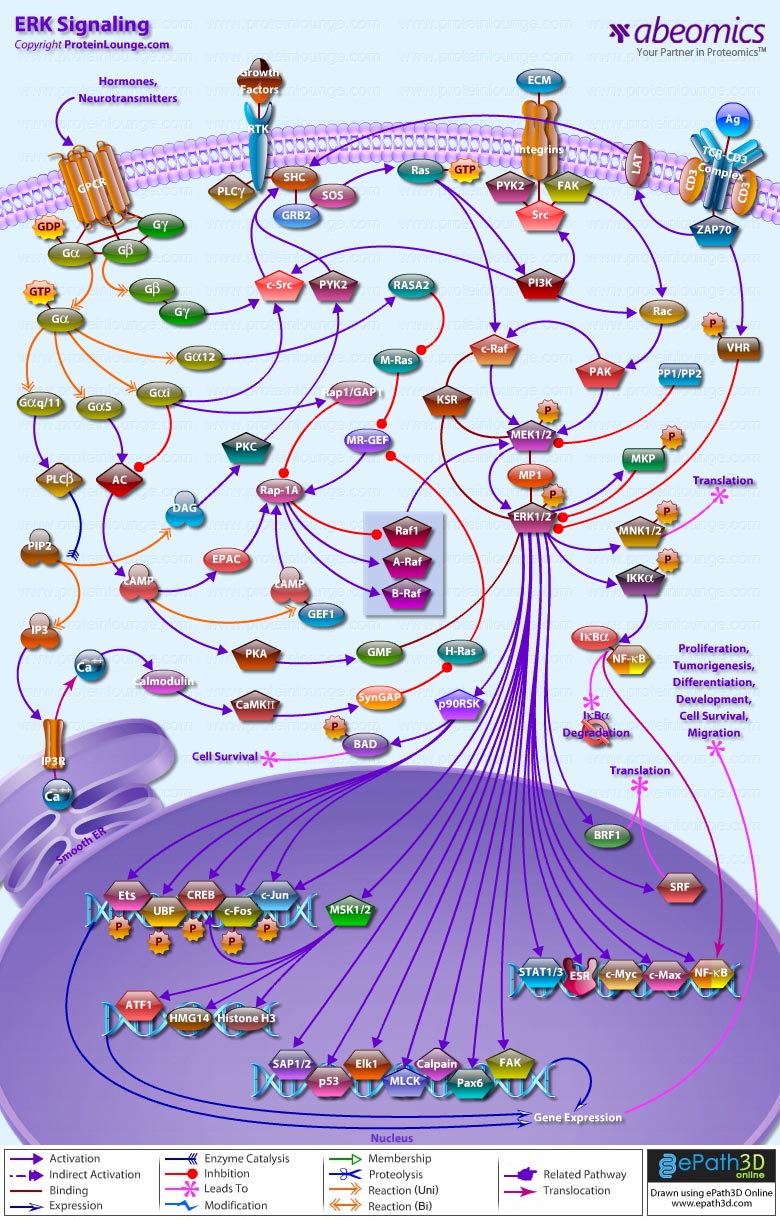
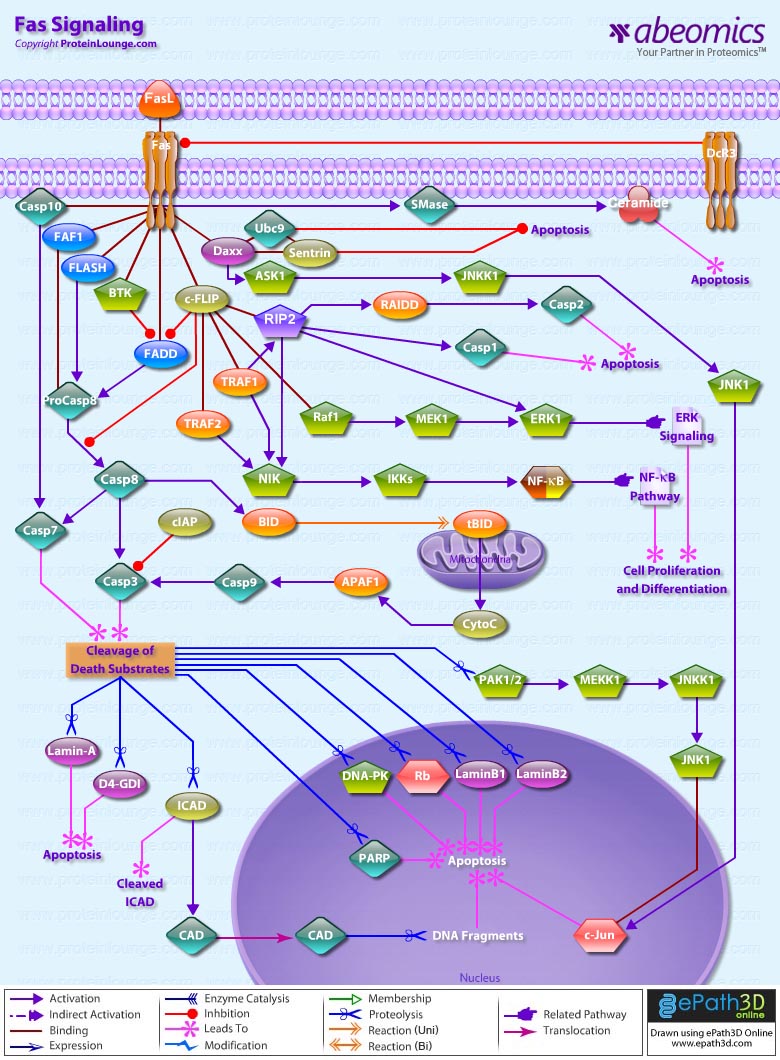
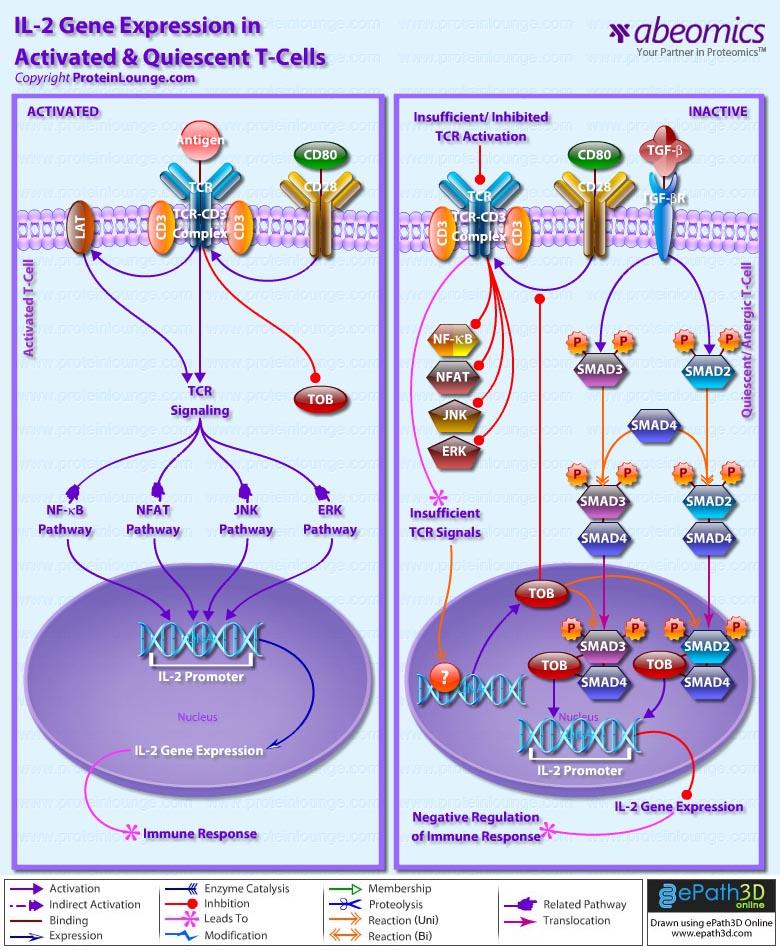
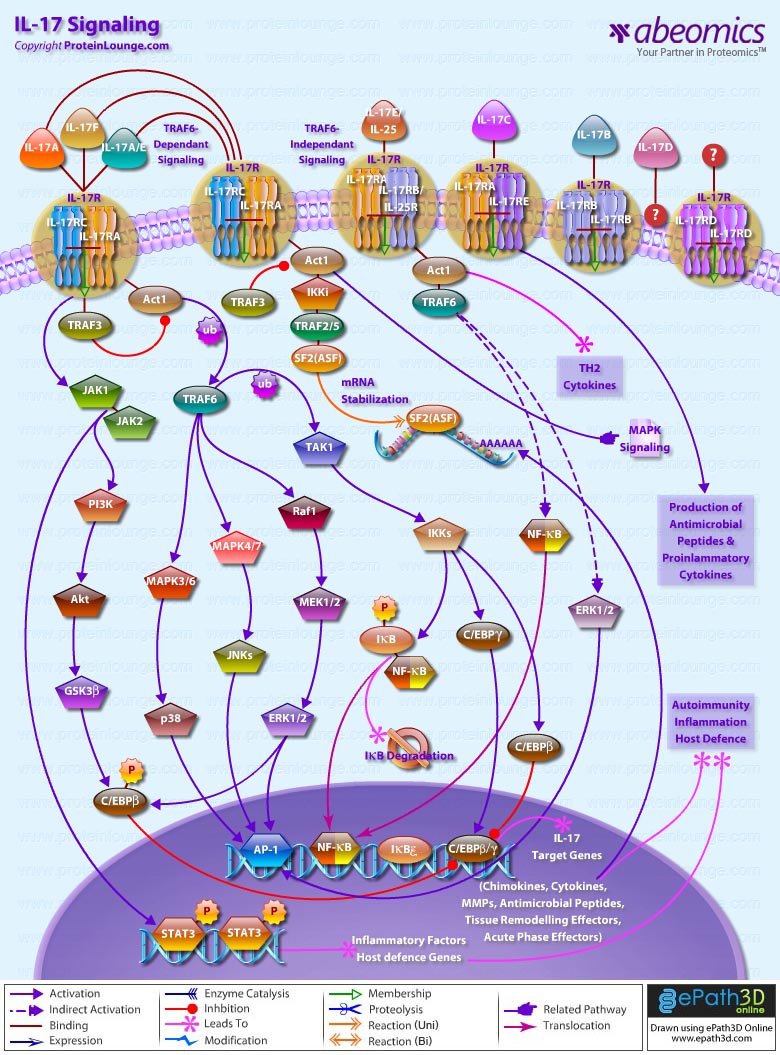
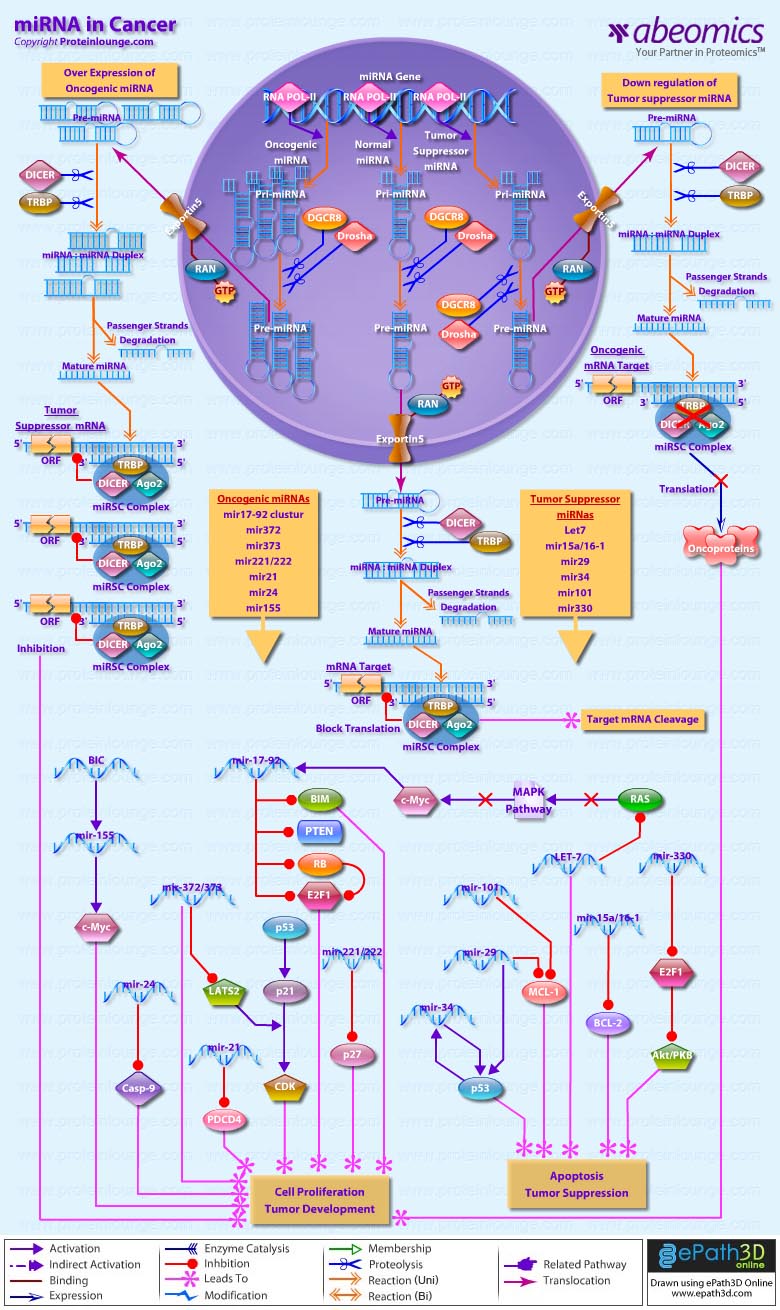
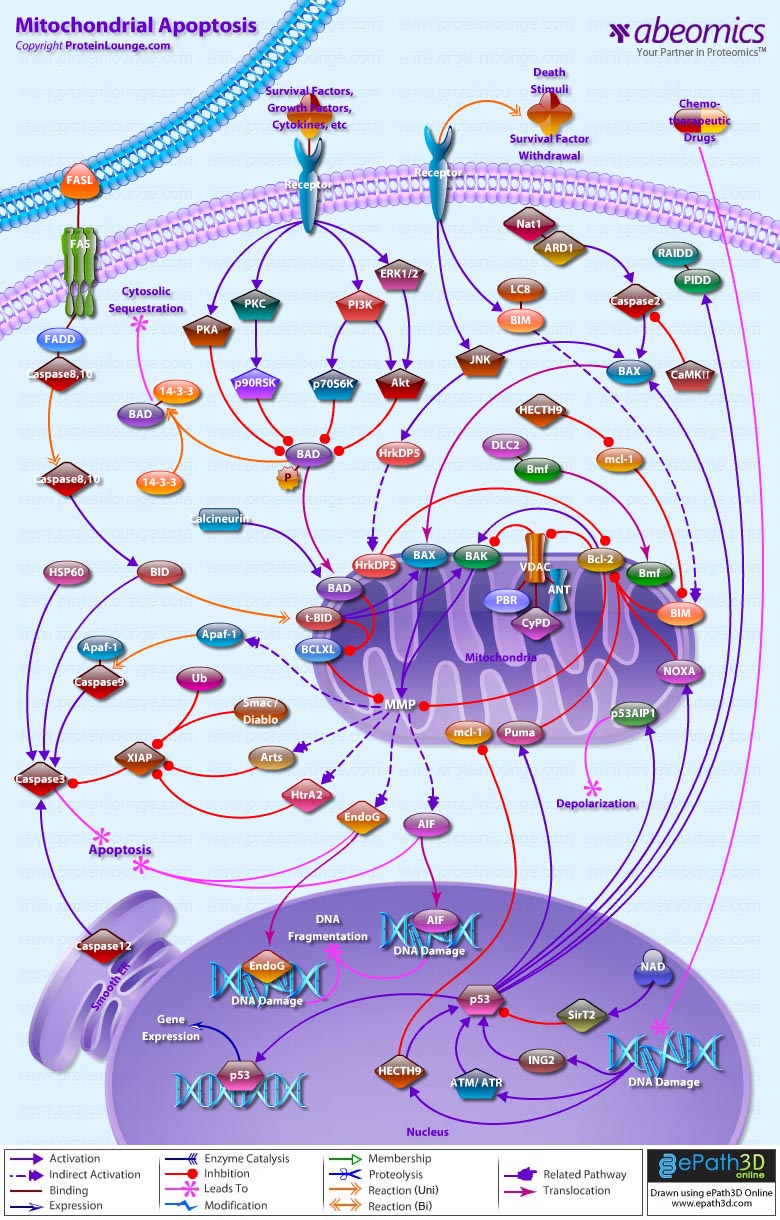
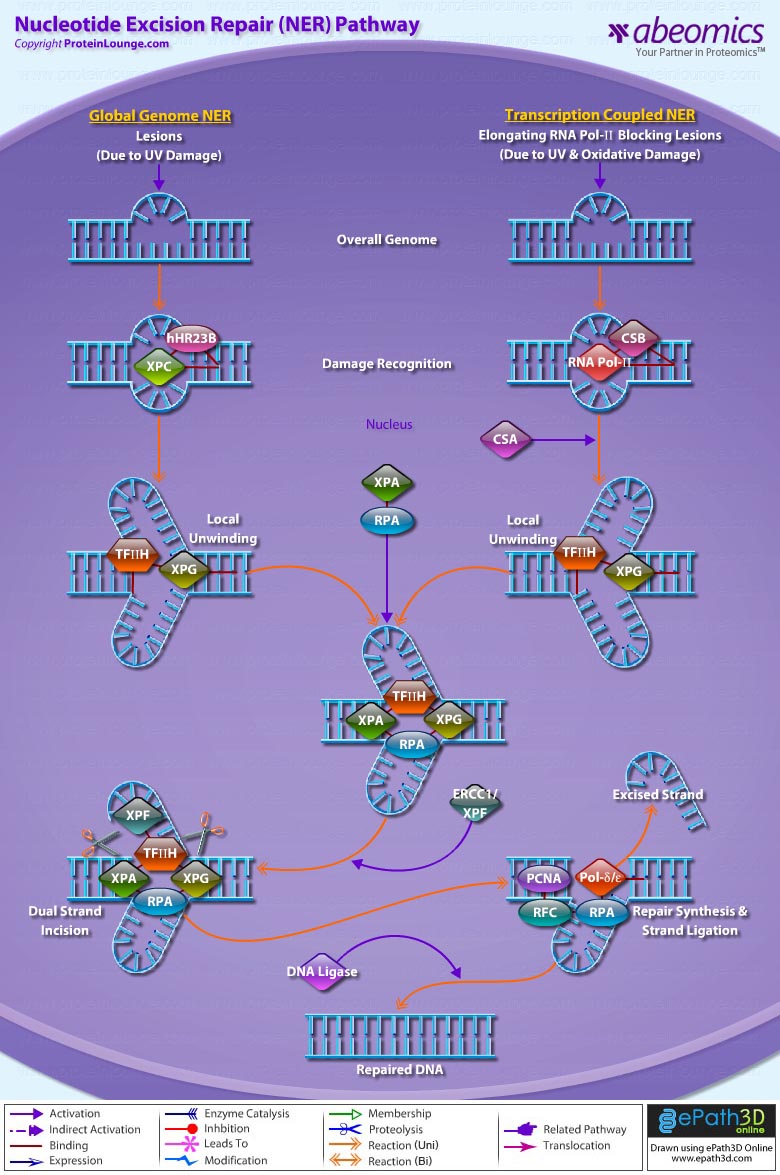
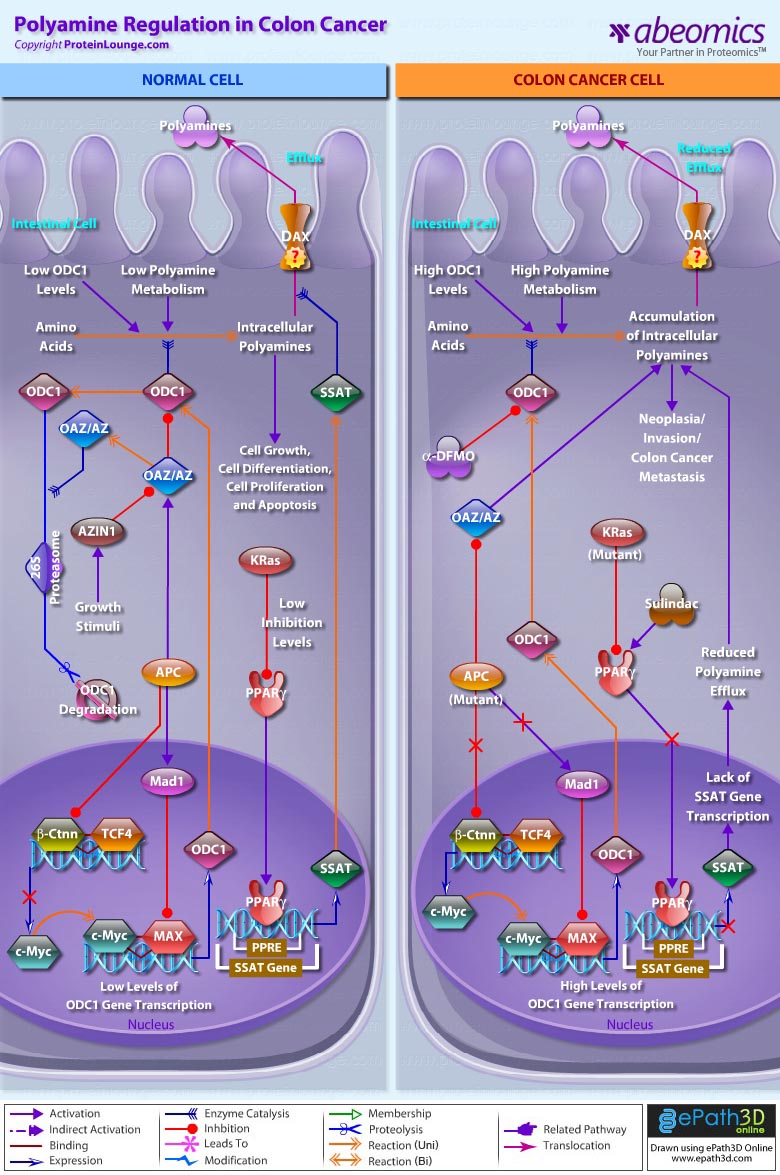
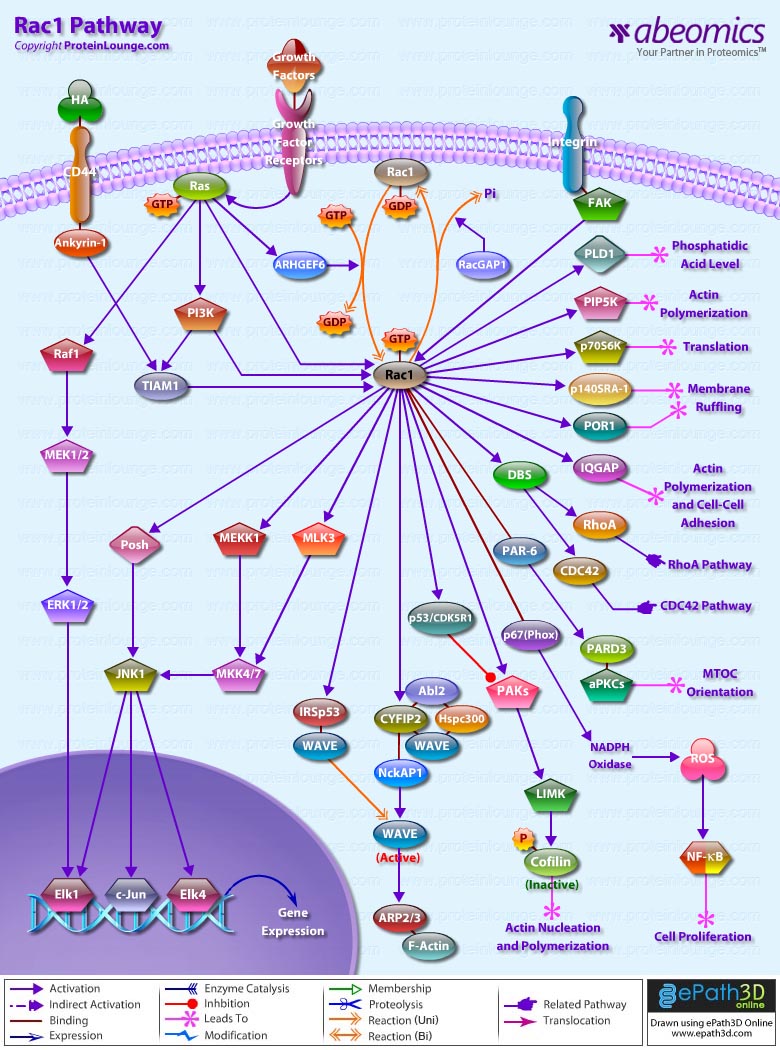
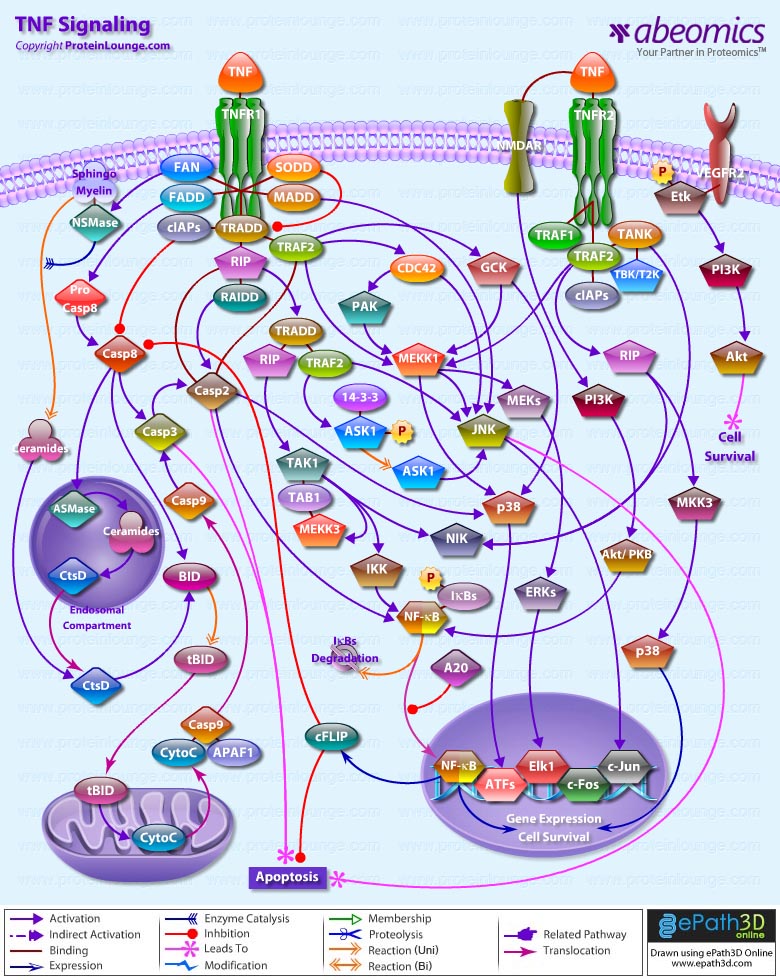
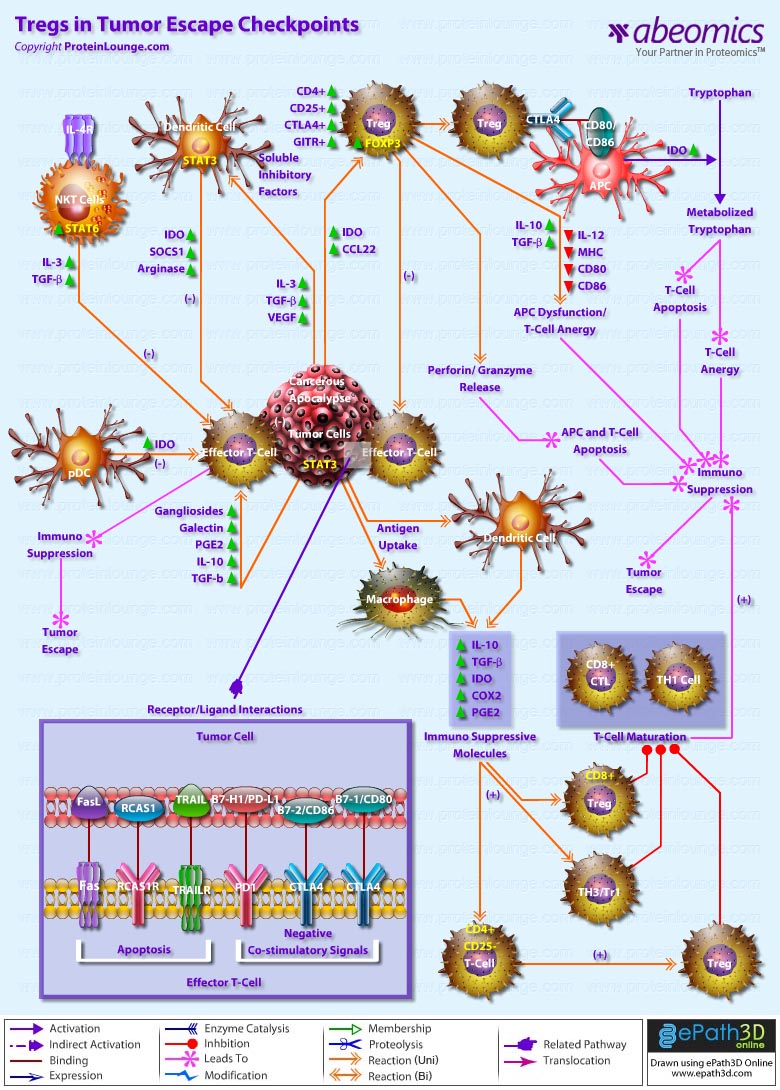
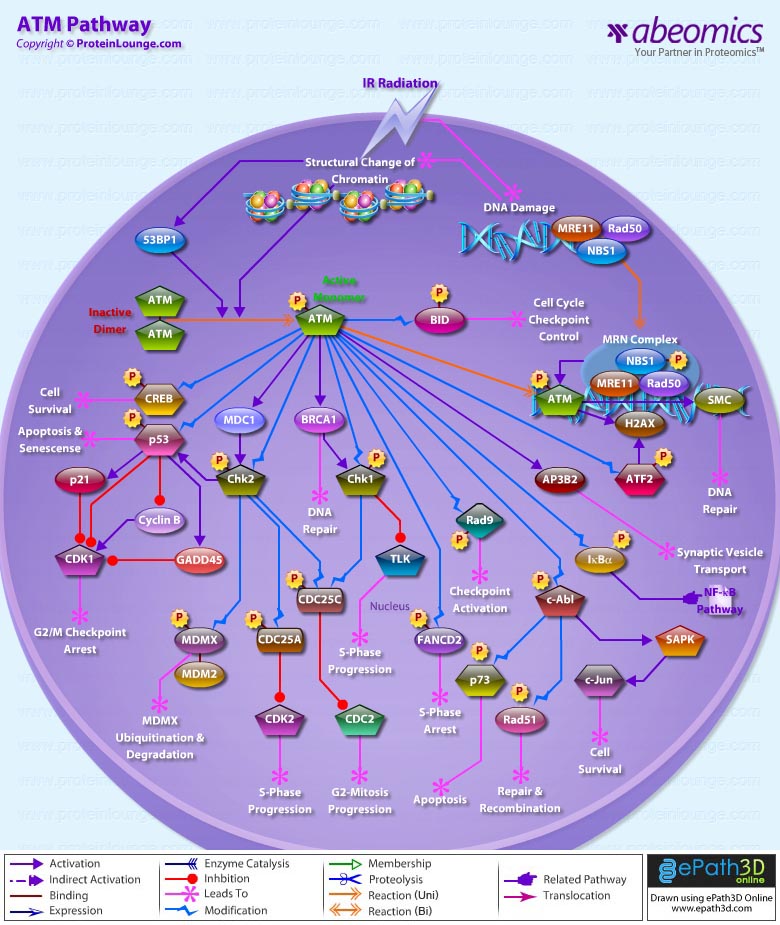
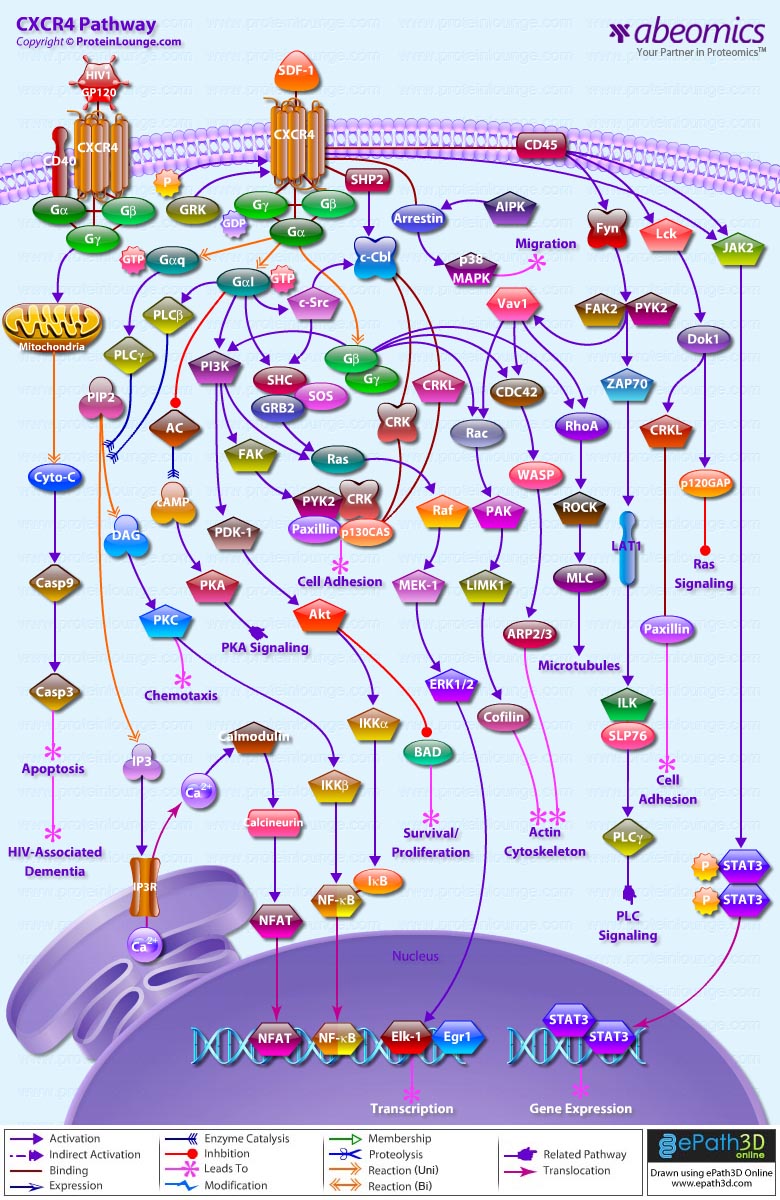
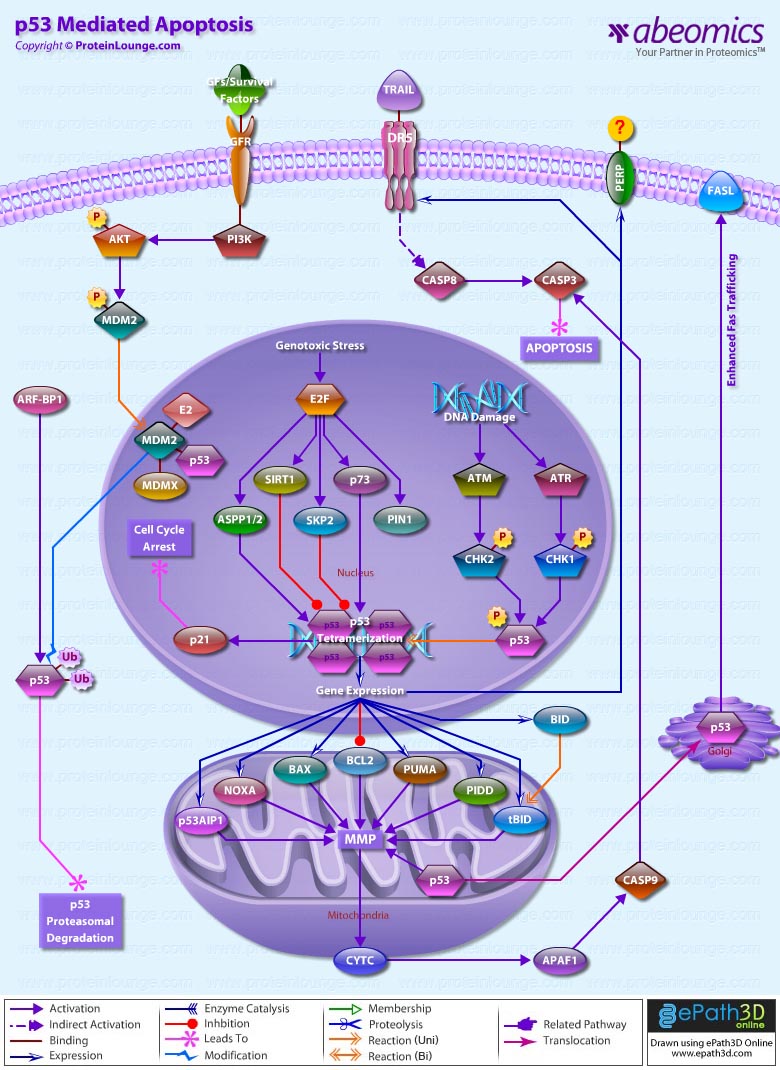
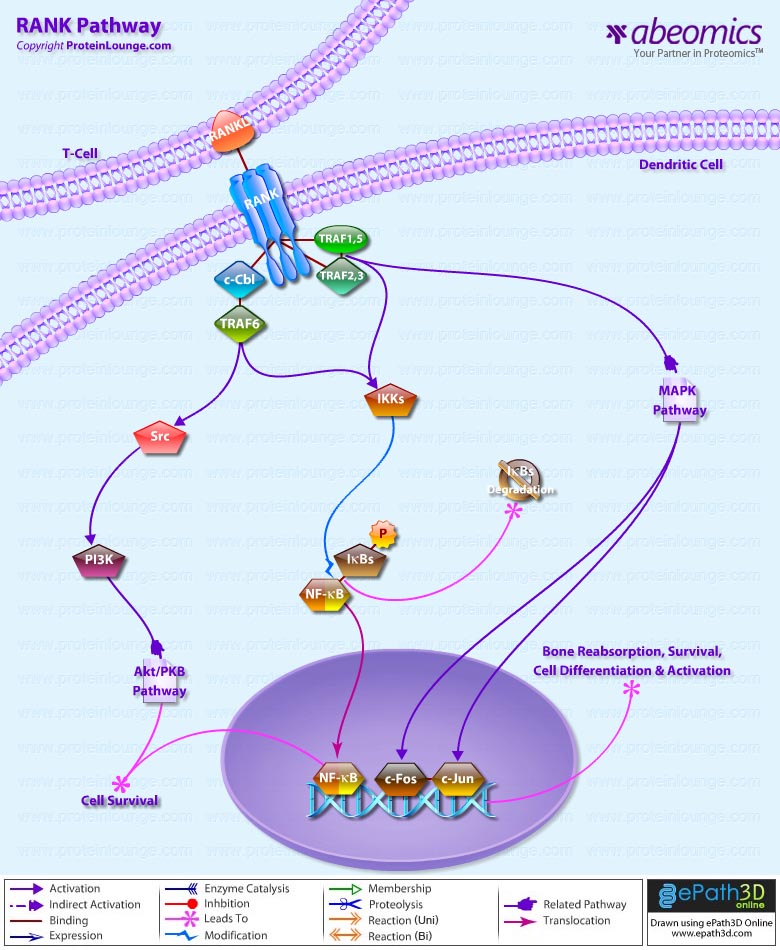
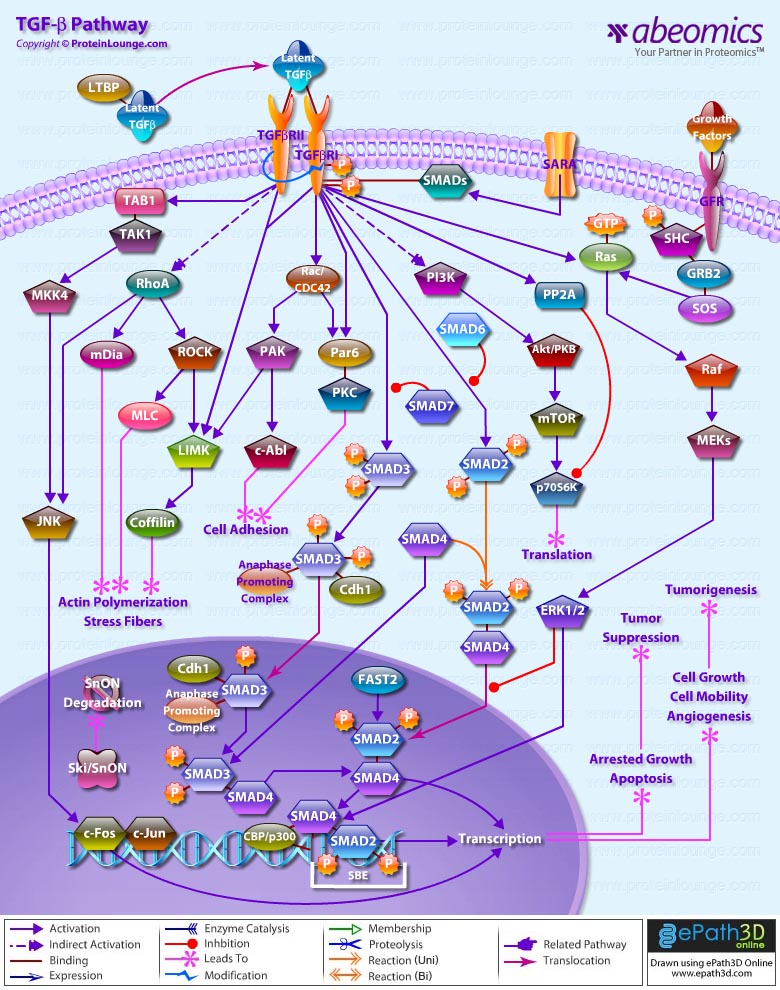
Stem cells are undifferentiated cells capable of producing virtually all cell types in our body. They are characterized by the ability to Self-renew and maintain Pluripotency. For proper developmental outcome, ESCs (Embryonic Stem Cells) must tightly regulate their differentiation status. Hundreds of genes have been identified, including several transcription factors, which have expression patterns tightly correlated with ESC differentiation. Very low number of transcription factors is responsible for regulating the development of an entire organism. These transcription factors form multiprotein complexes on DNA, thereby orchestrating the correct temporal–spatial expression of developmental genes. The process leads to the establishment of functional partnerships, with the combination rather than the individual activity of each factor eliciting specific transcriptional outcomes. Two key transcription factors, Oct4 (Octamer Binding Transcription Factor-4) and Nanog, have been identified that are crucial for maintenance of the Pluripotent state of ESCs (Ref.1 & 2).
Oct4, also known as Oct3, is a homeodomain transcription factor of the POU (Pit-Oct-Unc) family. The POU family of transcription factors can activate the expression of their target genes through binding an octameric sequence motif of an AGTCAAAT consensus sequence. Oct4 (POU5F1) plays a central role in Self-renewal, Pluripotency, and Lineage commitment in ESCs, the Embryonic Epiblast and PGCs (Primordial Germ Cells). Oct4 expression levels should be strictly maintained to regulate cell fate. Alterations in Oct4 expression promote differentiation and leads to the specification of Ectodermal, Endodermal, or Mesodermal primitive progenitors. Oct4 Protein consists of 3 domains: N-terminal domain, POU domain and a C-Terminal domain. POU domain consists of two structurally independent subdomains: a 75 amino acid amino-terminal POU specific (POUs) region and a 60 amino-acid carboxyl-terminal homeodomain (POUh). Both domains make specific contact with DNA through a helix-turn helix structure and are connected by a variable linker of 15 to 56 amino-acids. Regions outside the POU domain are not critical for DNA binding and exhibit little sequence conservation. The N-terminal domain (N-domain) is rich in Proline and Acidic residues, while the C-terminal domain (C-domain) is rich in Proline, Serine and Threonine residues. The N-domain plays an important role in transactivation. The C-domain also plays a role in transactivation. The activity of Oct4 C-domain is cell type specific and is regulated through phosphorylation, whereas the N-domain is not. Oct4 POU-domain functions differently by serving as interaction sites for cell type-specific regulatory factors (Ref.3 & 4).
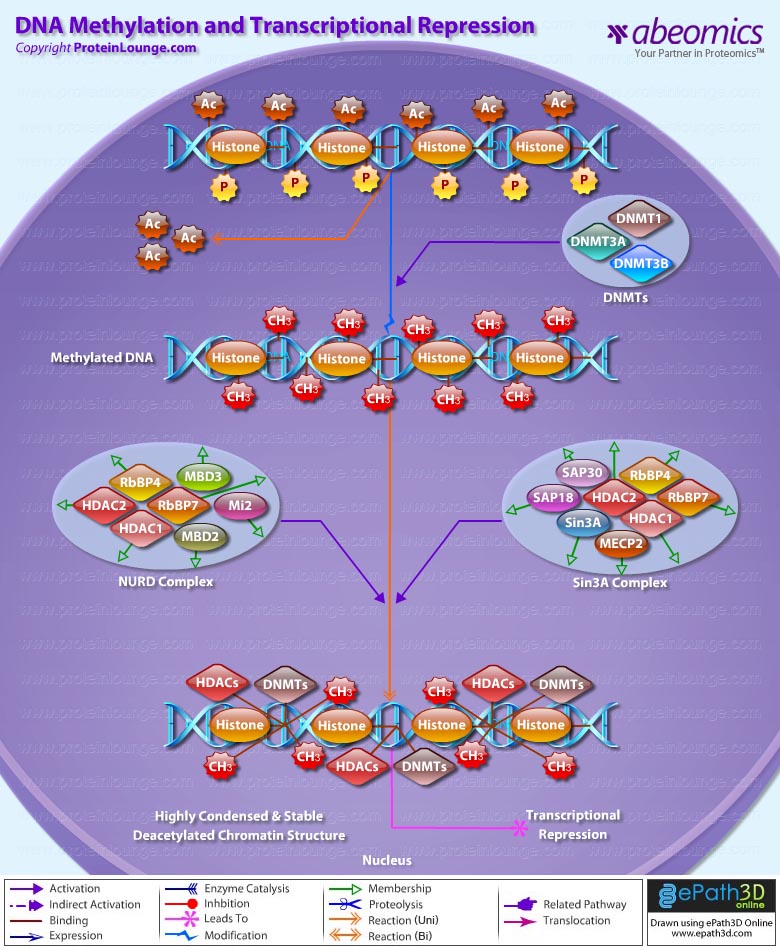
The precise mechanisms by which the activation and expression of Oct4 genes are regulated still remain to be elucidated. To date, two regulatory elements, a Distal and a Proximal Enhancer, have been identified as stem-cell-specific enhancers of Oct3/4 gene, to which many positive and negative regulators are recruited. There are 4 regions that are highly conserved among Human, Bovine and Mouse Oct4 promoter/enhancer elements, designated as CR1 (Conserved Region-1), CR2, CR3 and CR4 respectively. CR1 is downstream of Proximal Enhancer and immediately upstream of Exon 1. Each enhancer contains multiple potential binding sites for transcription factors that can either activate or repress Oct4 expression. In addition, methylation in these regions represses Oct4 expression in differentiated cells. Several positive and negative regulators bind to the Oct4 gene to regulate its expression. Among them, members of the orphan nuclear receptor superfamily, which can bind to the Proximal Enhancer, are known to influence Oct3/4 expression. LRH-1 (Liver Receptor Homolog-1), also known as NR5A2, is a putative positive regulator of Oct3/4. Other positive regulators include SF1 (Steroidogenic Factor-1) and RXR-Beta (Retinoid X Receptor-Beta). By contrast, GCNF (Germ Cell Nuclear Factor) or NR6A1 is a potential Oct3/4 negative regulator. COUF-TFI/II (Chicken Ovalbumin Upstream promoter-Transcription Factors- I/II), encoded by NR2F1 and NR2F2, respectively, also function as negative regulators of Oct3/4 expression. The balance between these positive and negative regulators might determine the precise level of Oct3/4 expression in response to extracellular stimuli. Oct4 can also be degraded by interacting with a Murine E3 Ub ligaseWwp2 (WW domain containing E3 Ubiquitin Protein ligase-2). Wwp2 can directly bind to Oct4 through WW domains. It promotes ubiquitination of Oct4 both in vitro and in vivo (Ref.1, 5 & 6).
Oct4 protein, once activated can act as an activator or repressor of target genes to maintain stem cell pluripotency. Oct4 usually functions in concert with other regulators to activate specific target genes in specific cell types at defined developmental stages. Most common regulators that cooperate with Oct4 in activating Oct3/4 target genes include SOX2 (SRY (Sex Determining Region-Y) Box-2) and Nanog. SOX2 is an HMG domain-containing transcription factor that binds to the consensus Motif CATTGTT. Oct4 and SOX2 bind to a few thousand regulatory sites in the ESC genome. They reciprocally regulate POU5F1 and SOX2 transcription via the Oct4-SOX2 complex in ESCs. In addition, Oct4 and SOX2 positively regulate Nanog, revealing that a tight transcriptional network is at work to maintain the undifferentiated state of ESCs. Besides, other ESC-specific enhancers that contain binding sites for Oct3/4 and SOX2 have been identified in several genes, including FGF4 (Fibroblast Growth Factor-4), SPP1 (Secreted Phosphoprotein-1), UTF1 (Undifferentiated embryonic cell Transcription Factor-1), FBXO15 (F-box Protein-15), and Lefty1 (Left right determination Factor-1) (Ref.1 & 7). Several other genes containing Oct4/SOX2 binding sites have recently been identified in Mouse. Genes containing 1 Oct4/SOX2 binding site include Lig3 (Ligase-III, DNA, ATP-dependent), KCTD3 (Potassium Channel Tetramerisation Domain containing-3), BIN1 (Bridging Integrator-1), Bmi1 (Bmi1 Polycomb ring finger oncogene) and NASP (Nuclear Autoantigenic Sperm Protein (histone-binding)). Genes containing 5 to 10 sites include InsIG2 (Insulin Induced Gene-2), Ipo11 (Importin-11), MYST4 (MYST Histone Acetyltransferase (monocytic leukemia)-4), NR6A1 (Nuclear Receptor Subfamily-6, group A, Member-1) and STRBP (Spermatid Perinuclear RNA Binding Protein). Genes with greater than 10 sites are POU2F1 (POU domain, class 2, Transcription Factor-1), RANBP17 (RAN Binding Protein-17), and SMYD3 (SET and MYND Domain containing-3). Thirty five genes implicated in chromatin remodeling are correlated to Oct4. Putative positive target genes include SWI/SNF members SMARCC1 (SWI/SNF related, Matrix associated, Actin dependent regulator of chromatin, Subfamily c, member-1), AT rich interactive domain (Swi1 like) containing proteins (ARID domains) ARID1A (AT Rich Interactive Domain-1A (SWI-like)), ARID5B (AT Rich Interactive Domain-5B (SWI-like)), JARID1B (Jumonji, AT Rich Interactive Domain-1B) and JARID2 (Jumonji, AT Rich Interactive Domain-2), which are direct Oct4 target. These ARID domain containing proteins, a subset of the Jumonji C family, have recently been associated with Histone Demethylase activity. Several other genes containing MYST, SET (SET translocation (Myeloid leukemia-associated)), and Chromo, and Bromo domains, which facilitate or recognize specific histone modifications, have also been identified. REST (RE1-Silencing Transcription factor) has been implicated in the repression of neuronal specific genes via its ability to recruit cofactors such as HDACs (Histone Deacetylases), CoREST (Co-repressor of Rest), Sin3, and MECP2 (Methyl CpG binding Protein-2). The identification of REST as a direct Oct4 target, in light of its role in maintaining chromatin plasticity throughout neurogenesis, provides a mechanistic understanding of Oct4’s role in promoting neural differentiation. CoREST and MECP2 are positively and negatively correlated to Oct4 respectively (Ref.8, 9 & 10).
Several members of the TrxG and PcG of transcription factors such as Ash1L (Ash1 (absent, small, or homeotic)-Like (Drosophila)), SUZ12 (Suppressor of Zeste-12 homolog (Drosophila)), ASH2L (Ash2 (absent, small, or homeotic)-Like (Drosophila)), Phc1 (Polyhomeotic-Like 1 (Drosophila)), and Rnf134 (Ring Finger protein-134), Bmi1 (BMI1 Polycomb ring Finger oncogene), and Phc3 (Polyhomeotic-Like 3 (Drosophila))are also correlated to Oct4, with the five latter genes validated as Oct4 targets. Diverse functions for PcG and TrxG genes in cancer, cell cycle control, and stem cell function have recently been described. The direct transcriptional regulation of several members of these complexes places Oct4 central to the coordination of these activities. The localization of SUZ12, a member of PRC2 (Polycomb Repressor Complex-2) at many Oct4 repressed loci in ESC provides indication that Oct4-Polycomb interaction may play a significant role in the active repression of lineage. Oct4 plays an important role in maintaining chromatin structure in Mouse ESC via regulation of and interaction with a unique constellation of PcG and TrxG complexes. While others are positive targets, Bmi1 and Phc3 are found to be negative targets of Oct4. About 38 cell-cycle related genes were positively correlated to Oct4 including CDC25A (Cell Division Cycle 25 homolog-A), GSPT1 (G1 to S Phase Transition-1), PPP1R8 (Protein Phosphatase-1, Regulatory (inhibitor) Subunit-8), CcnB2 (Cyclin-B2), CcnE1 (Cyclin-E1), CcnA2 (Cyclin-A2), CcnB1 (Cyclin-B1), and CcnF (Cyclin-F). Validated Oct4 target CcnF is implicated in cell cycle control at the G1/S and G2/ M checkpoints and has recently been associated with the maintenance of pRb (Retinoblastoma) in a hyperphosphorylated, inactive state. Conversely, the significantly Oct4 correlated PP1 (Protein Phosphatase-1) negative regulatory subunit Nipp1 (PPP1R8) may facilitate the functional inactivation of pRb. Other Cell cycle genes activated by OCT4 include D14Abb1E (DNA segment, Chr 14, Abbott 1 Expressed), IGF2BP1 (Insulin-like Growth Factor 2 mRNA Binding Protein-1) and JARID1B (Jumonji, AT Rich Interactive Domain-1B), which inactivate Rb. Inactivation of pRb is required for self-renewal. An imbalance in either of these processes, possibly emanating from deregulated signaling from the stem cell niche or mutations in the key regulators would lead to unrestrained cellular proliferation(Ref.11 & 12).
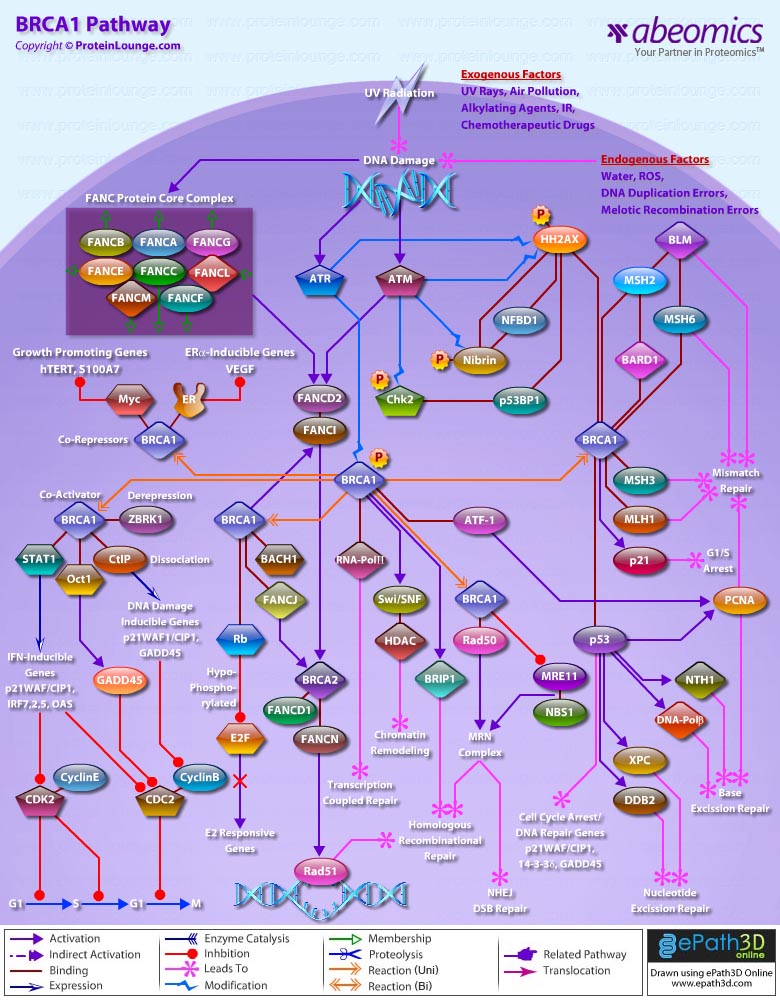
Twenty-five apoptotic genes are found to be positively correlated to Oct4, the majority of which,including AATF (Apoptosis Antagonizing Transcription Factor), API5 (Apoptosis Inhibitor-5), Aven (Apoptosis, caspase activation inhibitor), BAG4 (BCL2-Associated athanogene 4), Commd10 (COMM Domain containing-10), Nipa, and Opa1 (Optic Atrophy-1), function to inhibit Apoptosis. In addition, Bin1 (Bridging Integrator-1), Blp1 (BBP-Like Protein-1), Serpinb9 (Serpin peptidase inhibitor, Clade B (ovalbumin), member-9), SH3GLB1 (SH3-domain GRB2-like endophilin-B1), and Casp6 (Caspase6), all apoptosis inducing genes, were found to be negatively correlated to Oct4, with SH3GLB1 and Casp6 confirmed as targets. Thirty genes implicated in DNA damage and repair, are also positively correlated to Oct4. Members of the BASC (Brca1 Associated Surveillance Complex) including BRCA1 (Breast Cancer-1, early onset), Msh2 (MutS Homolog-2, colon cancer, nonpolyposis type 1 (E. coli)), MRE11A (MRE11 meiotic recombination 11 Homolog-A), Rad51 (RAD51 homolog (RecA homolog, E. coli), Blm (Bloom syndrome), Chek1 (CHK1 Checkpoint homolog), as well as PARP1 (Poly (ADP-ribose) polymerase family, member-1), TRp53 (Transformation Related Protein-53), FANCD2 (Fanconi Anemia, Complementation group D2), Tdrd7 (Tudor Domain Containing-7), and Xrcc5 (X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa)) are included. The validation of TRp53, Tdrd7, BRCA1, and PARP1 as direct Oct4 targets strengthens the importance of this group of genes in stem cell function. Tudor domain containing proteins (such as Tdrd7) play an important role in DNA damage response, Casp3 (Caspase3) in skeletal muscle differentiation, and PARP1, TRP53, and BRCA1 helps to modulate differentiation. Tdrd7 is a negative target of OCT4 (Ref.1 & 13).
PML (Promyelocytic Leukemia) and Coil (Cajal Bodies and PML Bodies), Gemin4 (Gem (nuclear organelle) associated Protein-4)and Gemin5 (Gem (nuclear organelle) associated Protein-5) (Gems), Nup35, Nup43, Nup54, Nup98, Nup133, Nup160, Nup188 (Nuclear Pore Complex), Ncl (Nucleolin) and Nolc1 (Nucleolus) and 46 genes implicated in RNA metabolism (Splicing Speckles, Spliceosome, Exosome, and Cajal Bodies) are positively correlated to Oct4. Several genes implicated in nuclear transport such as Ipo11, Kpna1 (Karyopherin Alpha-1 (Importin alpha 5)), Tnpo2 (Transportin-2 (Importin 3, karyopherin beta 2b)) and Tnpo3 (Transportin-3), Xpot (Exportin, tRNA (nuclear export receptor for tRNAs)), Gle1l (GLE1 RNA export mediator-like (yeast)), Xpo5 (Exportin-5) and Xpo6 and direct Oct4 targets IGF2BP1 and Phb were also positively correlated. Oct4 binding functions not only in the transcriptional repression of genes but also presents a means whereby these loci are organized spatially within the nucleus so as to be poised for activation given the appropriate cue. Oct4 promotes self-renewal by inactivating genes which promote differentiation or by activating genes which inhibit differentiation. Most common genes positively regulated by OCT4 include RAR-Alpha (Retinoic Acid Receptor-Alpha), REST, Aqr (Aquarius), JARID2 and SalL4 (Sal-like 4 (Drosophila)). RAR-Alpha, REST and Aqr inhibit Ectoderm and mesoderm differentiation. Negative targets of OCT4 include MEF2A (Myocyte Enhancer Factor-2A) and HoxB1 (Homeo Box-B1). In the initial stages of differentiation HoxB1 is transcriptionally activated which results in chromatin decondensation and reorientation of this locus to the nuclear centre. Finally, about 26 direct Oct4 transcriptional targets have been identified in Mouse ESCs (Ref.14).
In Human ESCs, Oct3/4 has also been reported to directly prevent differentiation towards trophectoderm by interacting with Cdx2 (Caudal type Homeobox-2), a trigger for trophectoderm differentiation, to form a repressor complex. This complex interferes with the autoregulation of these two factors, giving rise to a reciprocal inhibition system that establishes their mutually exclusive expression. As such, the downregulation of Oct3/4 results in an upregulation of Cdx2, and vice versa – a mechanism that might account for the two different pathways that lead to Pluripotent stem cells and to Trophectoderm cells. Oct4 represses gene expression either indirectly by neutralizing activators such as FoxD3 (Forkhead box-D3), or directly by binding to promoters. Oct4 repress the expression of FoxA1 and FoxA2 (Forkhead box-A2) through an interaction with the DNA binding domain of their activator FoxD3. Thus, Oct4 could prevent the differentiation of ESC lineages by acting like a co-repressor of lineage-specific transcription factors like FoxD3. Silencing of Ifn-Tau (Tau Interferon genes) appears to be mediated by Oct4. Oct4 and Ets2 (E26 avian leukemia oncogene 2, 3′ domain) form a complex through interaction between the Oct4 POU domain and the DNA binding domain of Ets2, and as a result quench the transactivation function of Ets2. Oct4 is downregulated, thus, alleviating the co-repression of Ets2 to allow the Trophoectoderm specific genes such as Ifn-Tau to be expressed. Oct4 also binds to a novel PSBP (Pluripotential cell-specific SOX Element-Binding Protein) leading to Nanog gene expression. OCT4 dominantly bound to the Octamer element, while PSBP preferentially bound to the SOX element (Ref.2 & 15).
Recent identification of Oct4 transcriptional targets in ESCs has revealed an unanticipated collaboration between Oct4, SOX2, and Nanog. The identification of common target sites in the regulatory elements of Oct3/4, SOX2 and Nanog has suggested that Oct3/4, SOX2 and Nanog might form a regulatory feedback circuit that maintains pluripotency in human and mouse ES cells; in this circuit, all three transcription factors regulate themselves, as well as each other. Although Oct4 maintain Pluripotency in combination with Nanog and SOX2, even a slight overdose of Oct3/4 triggers differentiation. Oct4 over-expression can inhibit its own expression as well as the expression of SOX2 and Nanog. Recently, a link between Oct4 and tumorigenesis has also been identified. Like ESCs, tumor cells exhibit a unique pattern of gene expression, and thus, may be under the control of one or more master regulators like Oct4. Therefore, an understanding of Oct4 function in Stem cell biology could also lead to novel treatments for certain malignancies (Ref.1 & 16).
References:
1.The human OCT-4 isoforms differ in their ability to confer self-renewal.
Lee J, Kim HK, Rho JY, Han YM, Kim J.
J Biol Chem. 2006 Nov 3;281(44):33554-65.
2.Self-renewal and differentiation of mouse embryonic stem cells as measured by Oct4 expression: the role of the cAMP/PKA pathway.
Faherty S, Fitzgerald A, Keohan M, Quinlan LR
In Vitro Cell Dev Biol Anim. 2007 Jan;43(1):37-47.
3.Oct4 targets regulatory nodes to modulate stem cell function.
Campbell PA, Perez-Iratxeta C, Andrade-Navarro MA, Rudnicki MA.
PLoS ONE. 2007 Jun 20;2(6):e553.
4.Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells.
Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J.
Stem Cells. 2007 Feb;25(2):500-10.
5.Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development.
Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ.
Mol Cell Biol. 2005 May;25(9):3492-505.
6.Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation.
Gu P, LeMenuet D, Chung AC, Mancini M, Wheeler DA, Cooney AJ.
Mol Cell Biol. 2005 Oct;25(19):8507-19.
7.Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation.
Wei F, Scholer HR, Atchison ML.
J Biol Chem. 2007 Jul 20;282(29):21551-60.
8.Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract.
Freberg CT, Dahl JA, Timoskainen S, Collas P.
Mol Biol Cell. 2007 May;18(5):1543-53.
9.Histone deacetylase activity is required for embryonic stem cell differentiation.
Lee JH, Hart SR, Skalnik DG.
Genesis. 2004 Jan;38(1):32-8.
10.Transplanted embryonic stem cells successfully survive, proliferate, and migrate to damaged regions of the mouse brain.
Srivastava AS, Shenouda S, Mishra R, Carrier E.
Stem Cells. 2006 Jul;24(7):1689-94. Epub 2006 Mar 30.
11.Identification of a nuclear localization signal in OCT4 and generation of a dominant negative mutant by its ablation.
Pan G, Qin B, Liu N, Scholer HR, Pei D.
J Biol Chem. 2004 Aug 27;279(35):37013-20.
12.Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers.
Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M.
Genes Dev. 2003 Aug 15;17(16):2048-59.
13.Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells.
Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F.
J Biol Chem. 2005 Feb 18;280(7):5307-17.
14.Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1.
Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B.
Nat Cell Biol. 2006 Oct;8(10):1114-23.
15.Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst.
Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J.
Development. 2005 May;132(9):2093-102.
16.Stimulation of Oct-4 activity by Ewing’s sarcoma protein.
Lee J, Rhee BK, Bae GY, Han YM, Kim J.
Stem Cells. 2005 Jun-Jul;23(6):738-51.

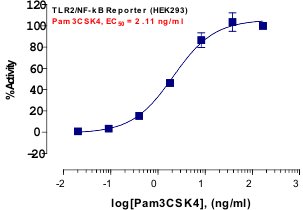
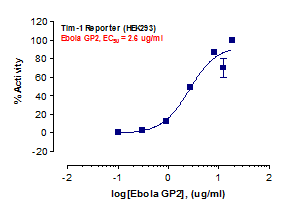
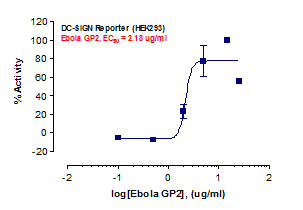
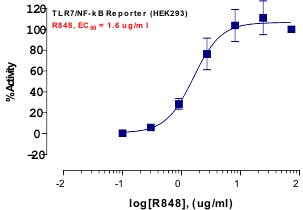
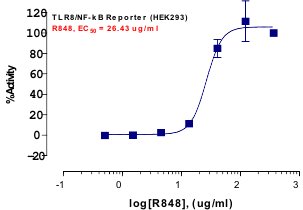
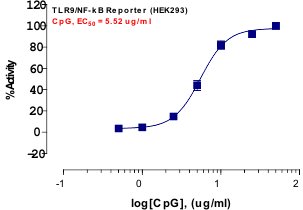
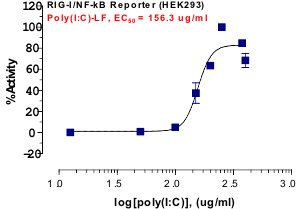
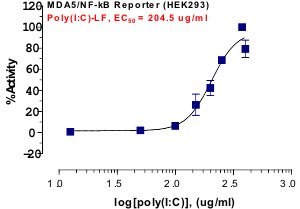
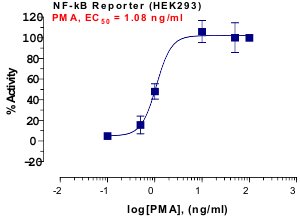
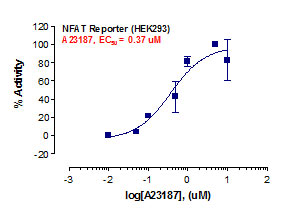
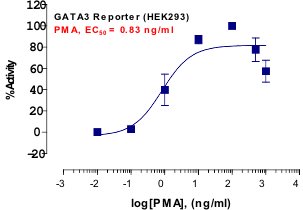
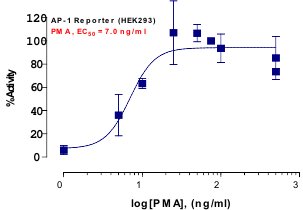
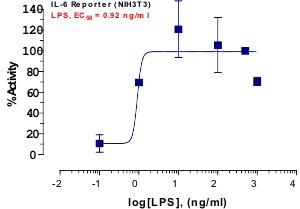
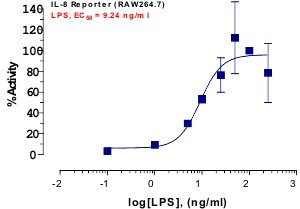
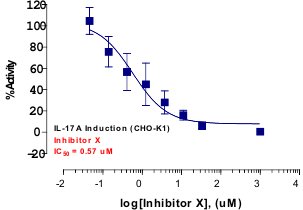
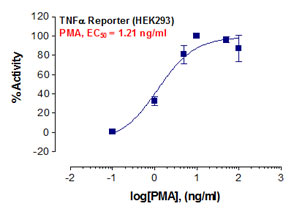
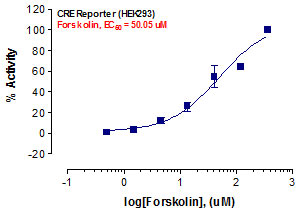
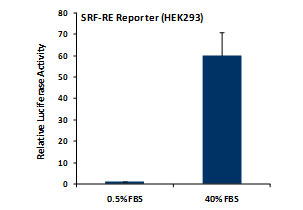
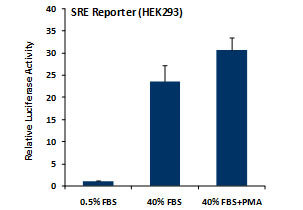
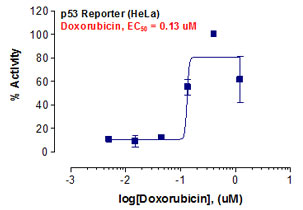
_Reporter_Assay.jpg)