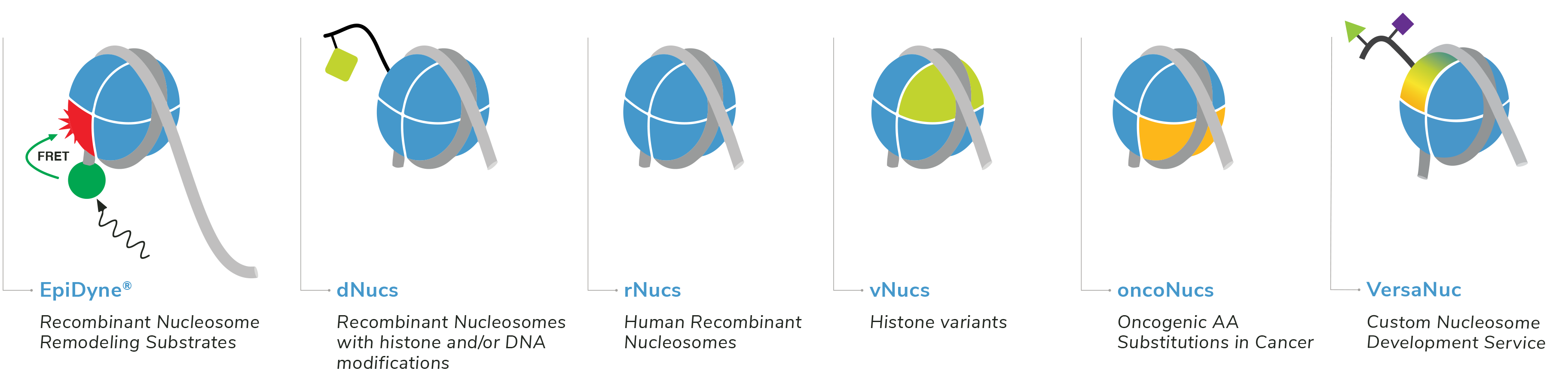
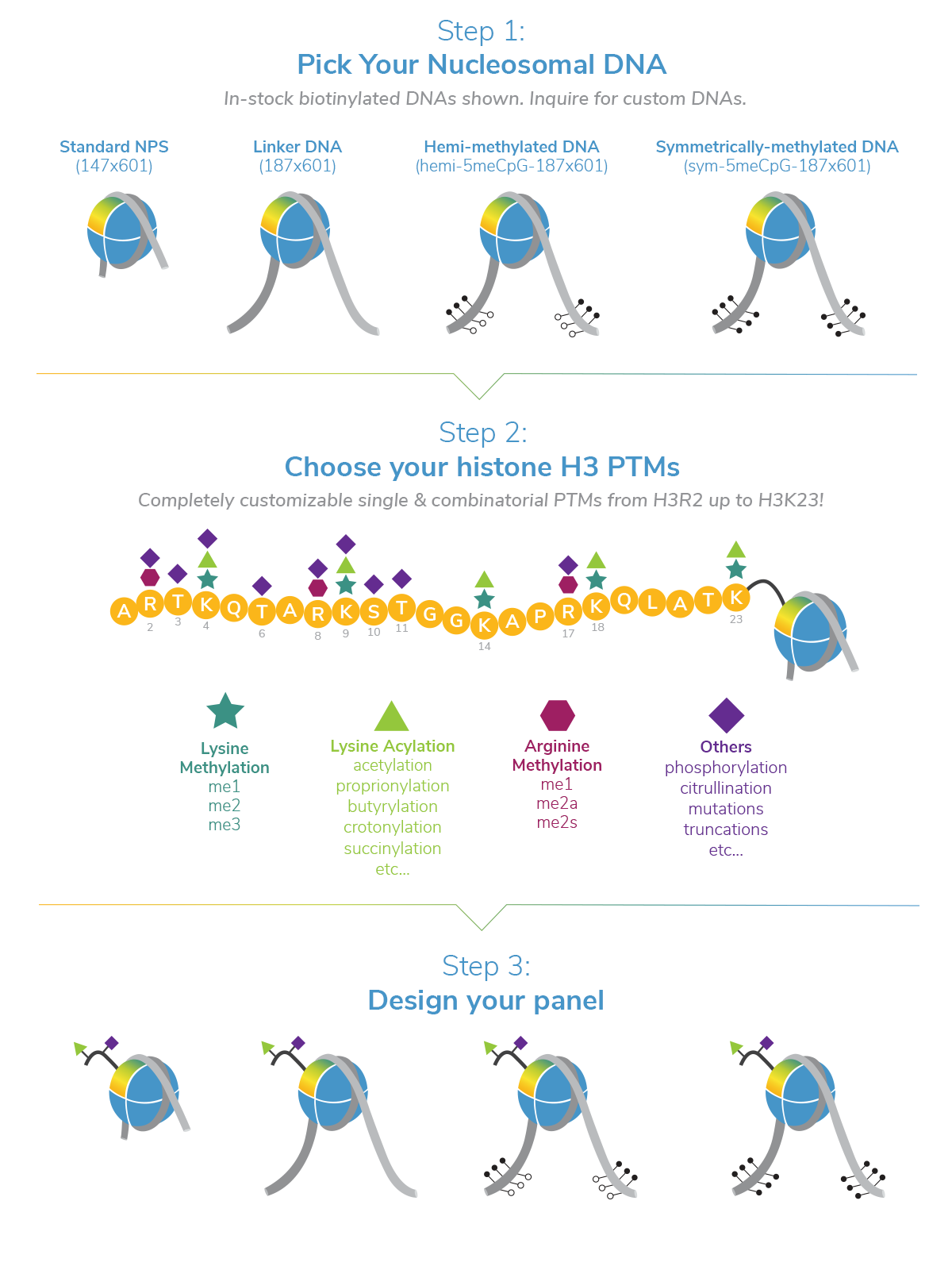
EpiCypher is dedicated to creating novel platforms that support innovative chromatin research. We are constantly expanding our product lines to provide the latest in quantitative chromatin technology.
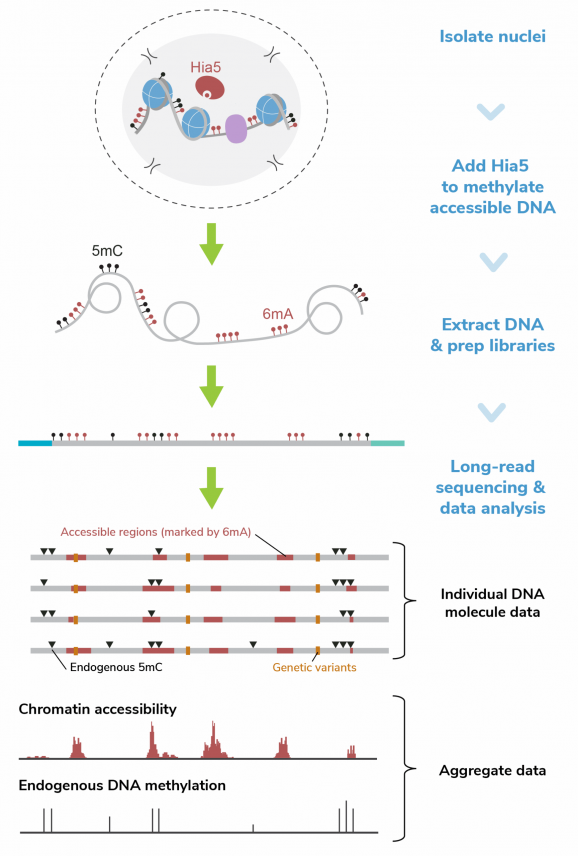
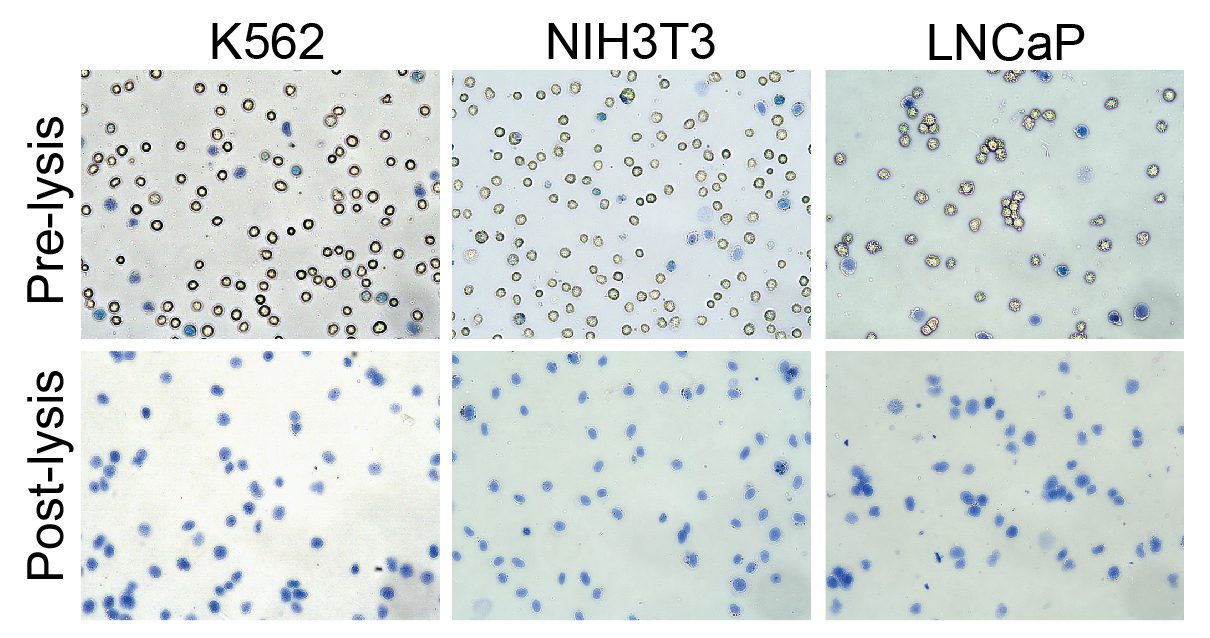
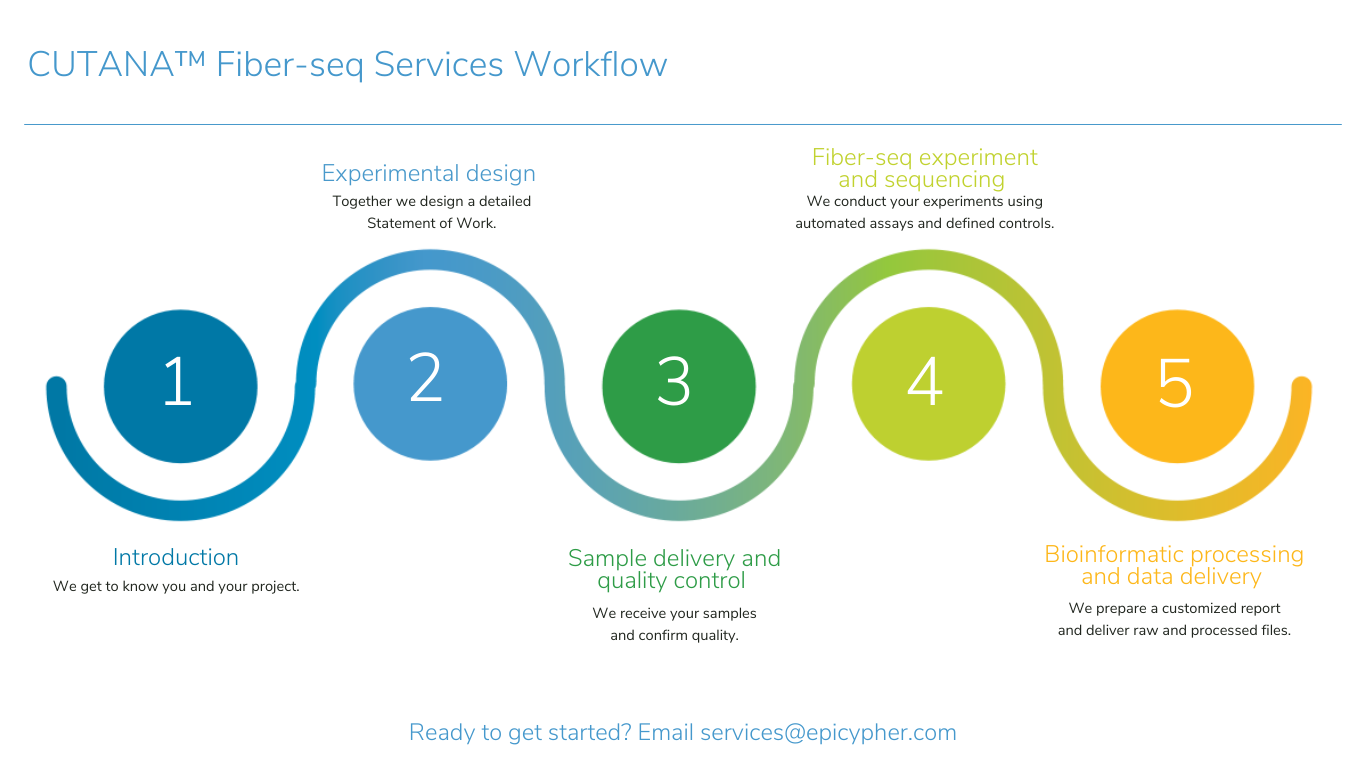
EpiCypher is leading the development of innovative research solutions to advance the science of epigenetic regulation and improve human health. As part of this drive, EpiCypher developed CUTANA™ CUT&RUN and CUT&Tag technologies as scalable, affordable, and ultra-sensitive alternatives to ChIP-seq for chromatin mapping. This technology, along with our comprehensive class of nucleosome substrates, provides the tools you need to take your project to the next level.