Researchers who regularly perform cell culture are likely familiar with the process of trypsinization. A trypsinization protocol is used for cell dissociation, in order to move cells from one environment to another. Usually cell dissociation is performed so that cells can be moved from the plate or flask in which they are grown to a new suspended medium, from which they can be separated and studied further. In this process, a trypsin EDTA solution is applied to the cells. Trypsin EDTA solutions contain both the trypsin enzyme, which breaks down proteins on the cell surface in order to dissociate them from the container and each other, and EDTA, which allows trypsin to break down specific bonds in order to achieve this cell dissociation. The trypsin enzyme is a digestive enzyme found in mammals. When using a trypsin enzyme in a trypsin EDTA solution, however, “cell surface proteins are often cleaved, which leads to dysregulation of the cell functions.”1
Another issue with trypsinization can be the time lag between taking cells out of the incubator and the addition of lysis buffer for downstream protein and RNA studies. This time window is usually around 15 to 20 minutes, during which changes in gene expression can happen. Changes in expression are possible even after only a few minutes and have already been described for some genes.2,3,4 These changes can have implications for downstream analysis and interpretation of RNA expression.
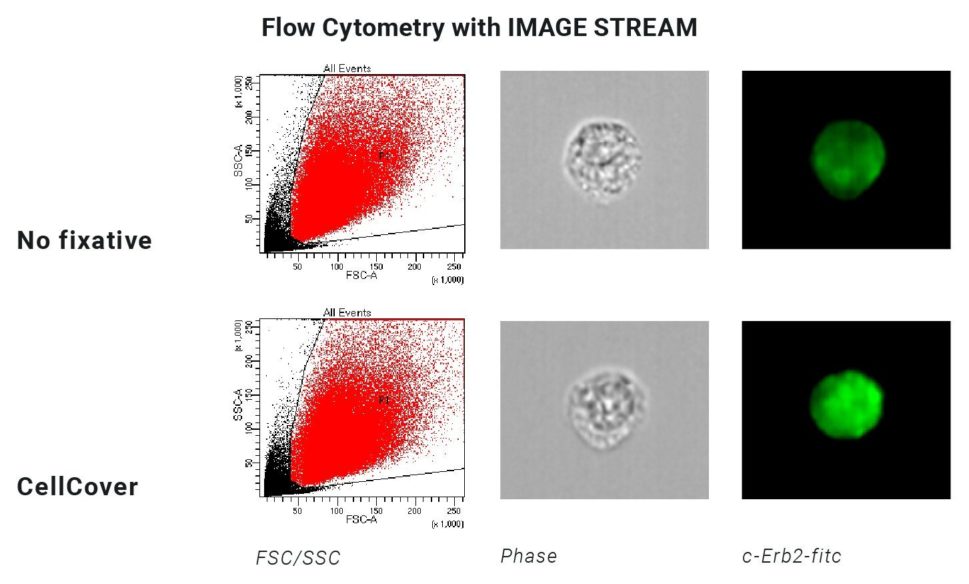
With a special protocol for CellCover this time gap is closed, which allows for the analysis of all proteins, RNA, DNA, and cell morphology at time point zero – the time at which cell samples are taken out of the incubator. With CellCover, the start of the trypsinization protocol and the time at which downstream analyses such as RNA sequencing happen are virtually indistinguishable.