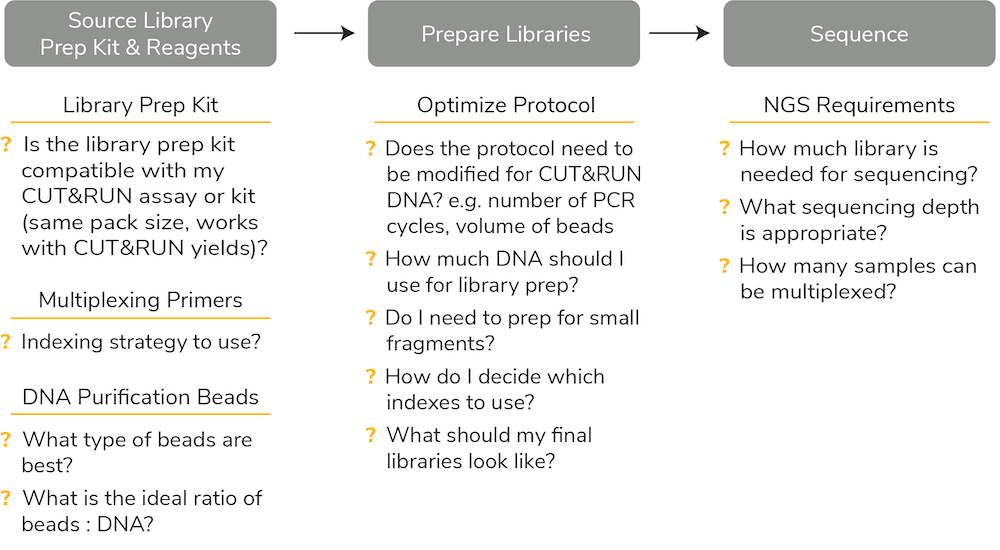
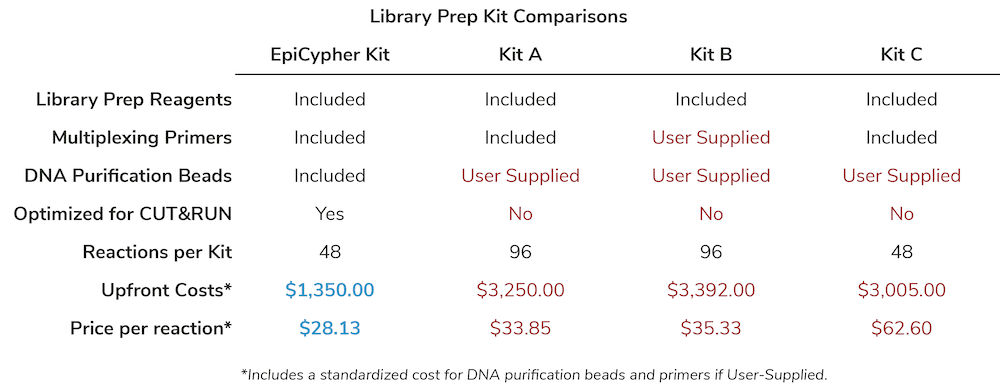
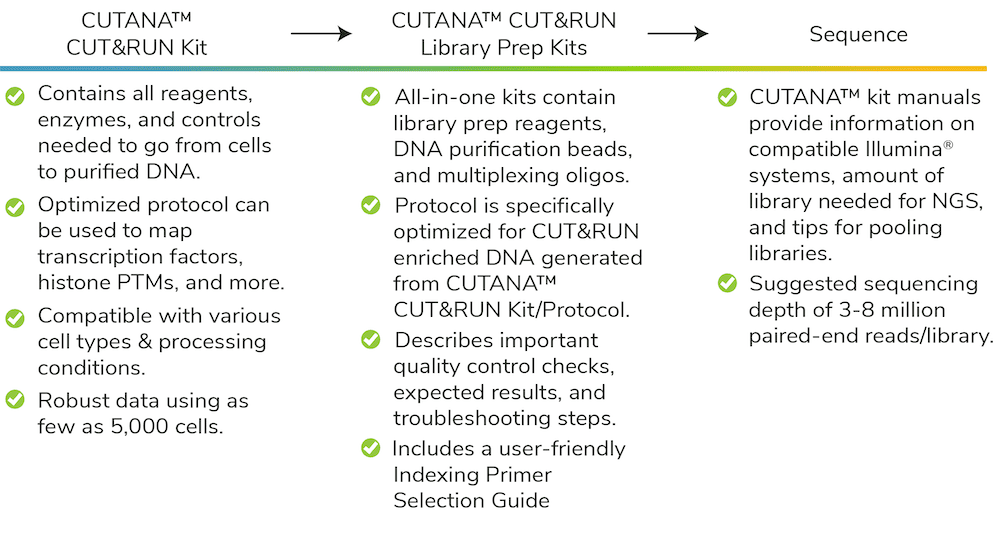
Figure 1: Historically, establishing a Library Prep plan for CUT&RUN required sourcing multiple reagents and optimizing existing protocols for your specific input.
Why are there so many decisions?
To maximize the application of their products, vendors offer general “DNA library prep kits” that can be combined with various multiplexing strategies and adapted for assay-specific inputs. These kits aren’t unique to CUT&RUN assays – in fact, few companies offer assay-specific library prep kits.
Despite the versatility of this product strategy, being “everything for everyone” has several drawbacks:
- The number of options makes it confusing to identify the best library prep method for your experiment.
- The generic nature of library prep protocols complicates integration with new profiling strategies, such as CUT&RUN.
- You have to spend extra money to source all the tools you need, since assay kits, library prep kits, DNA purification beads, and associated reagents come in different pack sizes from multiple vendors.
Can’t we just use ChIP-seq library prep protocols?
CUT&RUN users have relied on ChIP-seq library prep kits or generic DNA library prep kits adapted to lower DNA input. But modifying ChIP-seq library prep for ultra-sensitive CUT&RUN assays has been difficult for many reasons (see Figure 1):
- Lower yields from CUT&RUN challenges the use of ChIP-seq library prep protocols. Are lower inputs within the sensitivity limits of the library prep kit? Should the amount of library prep enzymes, sequencing adapter, or indexing primer be reduced? Are additional PCR cycles needed to ensure there is sufficient library for NGS?
- Unclear how to confirm high quality of CUT&RUN libraries. Fragment distribution analysis (e.g. by Agilent Bioanalyzer®) is an important quality control step during library prep, and data are used to pool samples for NGS. Libraries using CUT&RUN DNA appear different compared to ChIP-seq libraries, complicating this step.
- Lack of guidance on NGS requirements for CUT&RUN. One of the core advantages of CUT&RUN is high quality data at low sequencing depth. But many users continue to use 20 million (or more) reads per reaction, limiting multiplexing capabilities and increasing NGS costs.
The combination of decisions and assay optimization required for CUT&RUN library prep is intimidating, even for seasoned researchers, and has slowed the adoption of CUT&RUN by some groups.
EpiCypher decided to develop CUT&RUN assay-specific Library Prep Kits to provide a comprehensive solution to the concerns outlined above. The resulting CUTANA workflow makes it easier than ever to perform CUT&RUN assays, with major gains in throughput and reduced costs.
Now that we have covered “why” we made our Library Prep Kits, let’s dive into “how” we did it.