Bulk RNA barcoding and sequencing (BRB-seq) is an innovative, efficient, and cost-effective RNA sequencing technology that leverages the benefits method of early-stage barcoding and unique molecular identifiers (UMIs) to produce consistent, reproducible, and uniform RNA sequencing data (Alpern et al. 2019). Despite the utility of RNA sequencing (RNA-seq) in transcriptomics, traditional RNA-seq technologies are still too time consuming, costly, and analytically challenging to replace reverse transcription-quantitative real-time PCR (qRT-PCR) as the preferred method of quantifying changes in gene expression. In contrast, BRB-seq generates high-quality transcriptomic data with mere hours of hands-on time at a cost tantamount to profiling four individual genes using conventional qRT-PCR (Alpern et al. 2019).
Sample barcoding and early multiplexing first emerged in the field of single-cell transcriptomics as a way to reduce cost and handling time by generating a single sequencing library that contains multiple distinct samples (Ziegenhain et al. 2017). While other methods have attempted to address the shortcomings in bulk RNA sequencing, they tend to result in biased libraries or miss the mark on cost-effectiveness and handling time (Alpern et al. 2019). BRB-seq fills this previously unmet need by applying sample barcoding and early multiplexing principles to bulk RNA profiling, a highly optimized and rigorously evaluated process that matches the performance of far more costly methods and outperforms the best single-cell RNA-seq protocol, SCRB-seq (Alpern et al. 2019). Although BRB-seq does not allow for the analysis of full-length transcripts, splice variants, fusion genes, or address research questions involving RNA editing, it is arguably unnecessary in most cases to generate a sequencing library of complete transcripts.
The BRB-seq Innovation
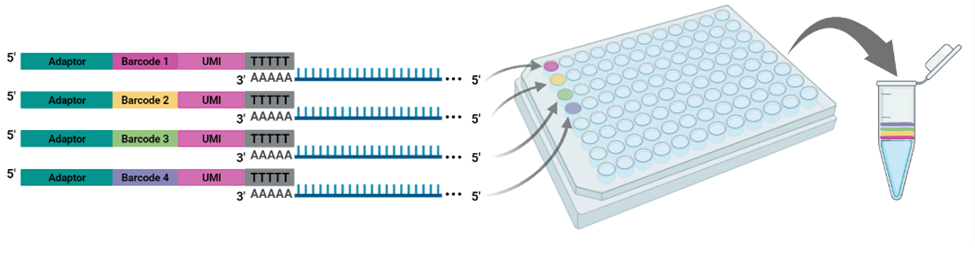
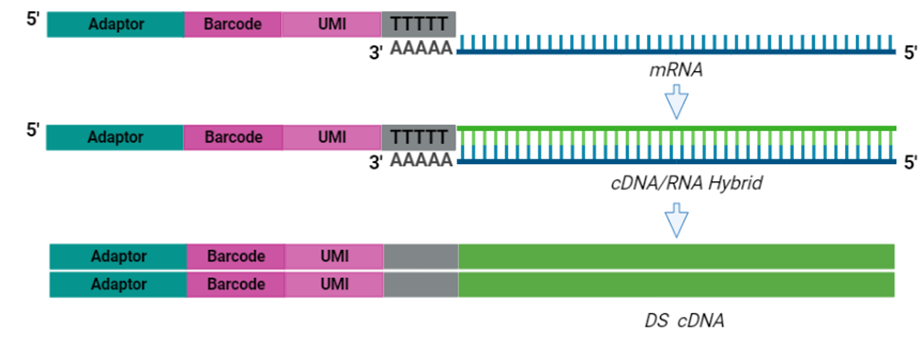
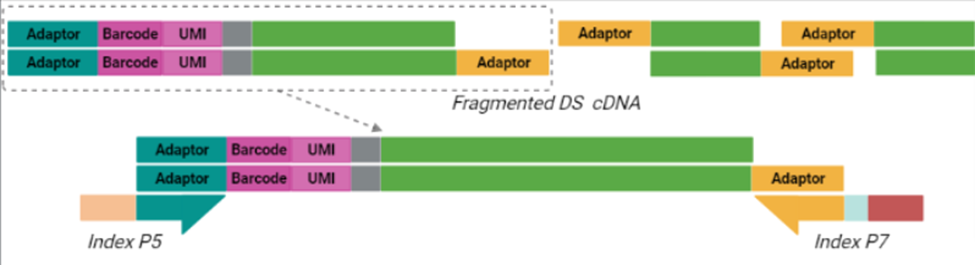
The cornerstone of BRB-seq technology is the use of highly optimized barcoded oligo(dT) primers that uniquely tag each individual RNA sample during the first-strand synthesis step of cDNA library preparation. The barcoded nucleotide sequence includes an adaptor for primer annealing, a 14-nt long barcode that assigns a unique identifier to each individual RNA sample, and a random 14-nt long UMI that tags each mRNA molecule with a unique identifier to distinguish between original mRNA transcripts and duplicates that result from amplification. Furthermore, BRB-seq allows individually barcoded RNA samples to be pooled, thereby streamlining subsequent steps in cDNA library preparation and sequencing (Figure 1), while the use of UMIs decreases amplification noise by eliminating PCR amplification bias (Alpern et al. 2019; Sena et al. 2018; Ziegenhain et al. 2017).