The largest blood cell genotyping and transcriptomic study ever performed has shown that a much higher number of common changes in the DNA sequence affect gene expression in blood cells than previously thought (Võsa et al. 2021). In this article, we look at how the use of RNA-seq in this study contributed to these exciting new discoveries in the genetic regulation of blood cells.
Summary:
Genetics is one of the deciding factors of traits and disease susceptibility. When changes in DNA sequence can explain variation in gene expression levels for a specific trait or disease, they are called expression quantitative trait loci (eQTLs) (Nica and Dermitzakis 2013).
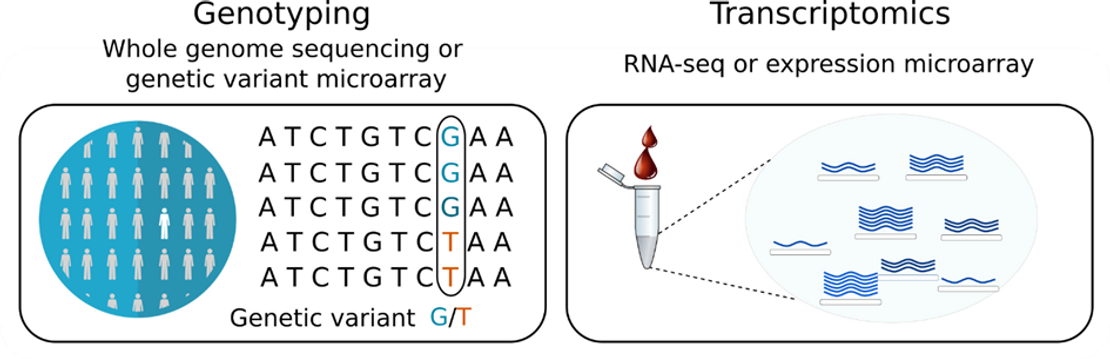
To detect eQTLs in blood cells, the eQTLGen Consortium analyzed a data set comprising high-throughput genomic and transcriptomic technologies, such as whole genome sequencing, RNA-seq and DNA variant and expression microarrays (Fig. 1) (Võsa et al. 2021). It included 37 studies with more than 31,000 study participants (Võsa et al. 2021).
Thanks to these high-throughput technologies including RNA-seq, and the large sample size, researchers discovered that genetic variants in close proximity to genes (cis-eQTLs) regulate 88% of all genes analyzed. This often includes a physical interaction between the variant and the gene (Võsa et al. 2021). In contrast, genomic variants over 5Mb away from genes (trans-eQTLs) regulate one third of all genes analyzed, likely due to transcription factor activity (Võsa et al. 2021). For example, one genetic variant possibly regulates the neuronal repressor REST and the expression of 88 neuronal genes in trans as a result (Võsa et al. 2021).
How RNA-seq was used:
eQTL detection relies firstly on genotyping to find genetic variations, followed by RNA-seq or microarray driven transcriptomics to detect gene expression variation for the whole genome of each individual (Fig. 1) (Nica and Dermitzakis 2013). Discovery of eQTLs are then made through the statistical association of these genetic variants with the expression level of the gene of interest. When the sample size is large enough, these statistics can even detect weak effects of genetic variants on gene expression.