Novel sequencing technologies based on the 3′ barcoding of mRNA now enable transcriptomic studies with higher sample numbers and lower costs than ever before.
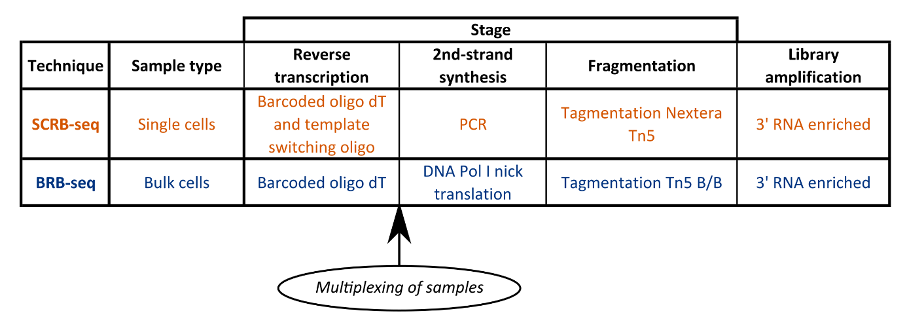
In this article we compare two of these techniques; Single Cell RNA Barcoding and sequencing (SCRB-seq) and Bulk RNA Barcoding and sequencing (BRB-seq) (Soumillon et al., 2014; Alpern et al., 2019).
What is SCRB-seq?
SCRB-seq is a high throughput technology for the mRNA sequencing of large numbers of single cells. It is reliant on the early barcoding and subsequent multiplexing of samples (Soumillon et al., 2014).
Single cells are isolated into wells and lysed. mRNA transcripts are then annealed to custom primers comprising a poly(T) tract, unique molecular identifier (UMI) and well barcode. Template switching reverse transcription and PCR amplification generates full length, barcoded cDNA.
Because mRNA from each cell (one per well) has a unique barcode, researchers can pool all samples and identify each after sequencing. They can also use the UMI to detect PCR duplicates.
Libraries are then prepared for the multiplexed samples using the Nextera XT library prep with modified i5 primers, followed by sequencing.
The creators of SCRB-seq used it to profile over 12,000 cells throughout the development of human fat cells revealing major differences in gene expression between different cell types and time points (Soumillon et al., 2014).
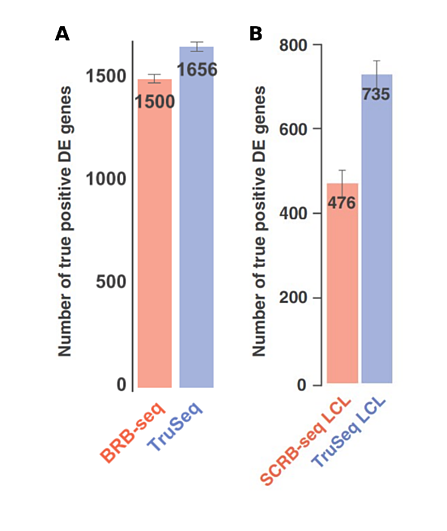
Some studies have used unmodified SCRB-seq for bulk samples, however Alpern et al. optimized BRB-seq specifically for this purpose, with superior results (Cacchiarelli et al., 2015; Kilens et al., 2018; Alpern et al., 2019).
How is the BRB-seq workflow different from SCRB-seq?
Like SCRB-seq, the primer barcode used in BRB-seq includes a poly(T) tract, UMI and well barcode, but the workflow differs from SCRB-seq in several key ways (Fig. 1) (Alpern et al., 2019).