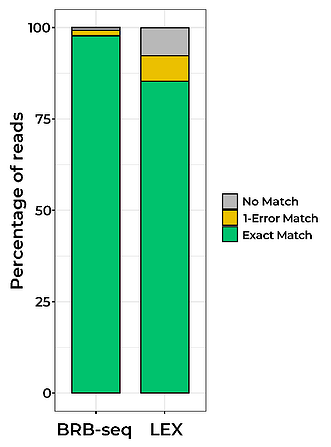
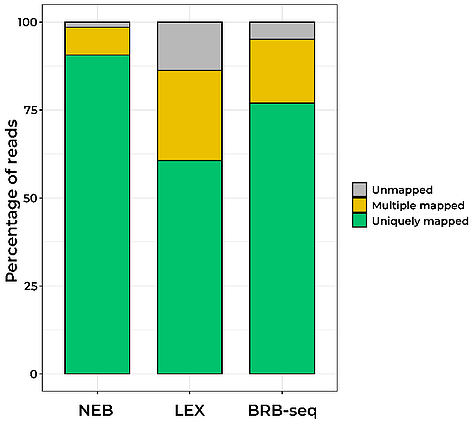
Since the initial BRB-seq publication (Alpern et al., 2019), we have continually optimized the BRB-seq workflow to lower the detection limit, reduce cross-contamination between samples, and expand the range of tissues and organisms where BRB-seq can be used. In particular, our novel globin depletion kit now allows efficient sequencing of blood samples.
Given these improvements, in this article we compare our optimized BRB-seq protocol to two widely used state-of-the-art library preparation technologies; the QuantSeq-Pool (LEX) and NEBNex Ultra II (NEB).
Original BRB-seq comparison:
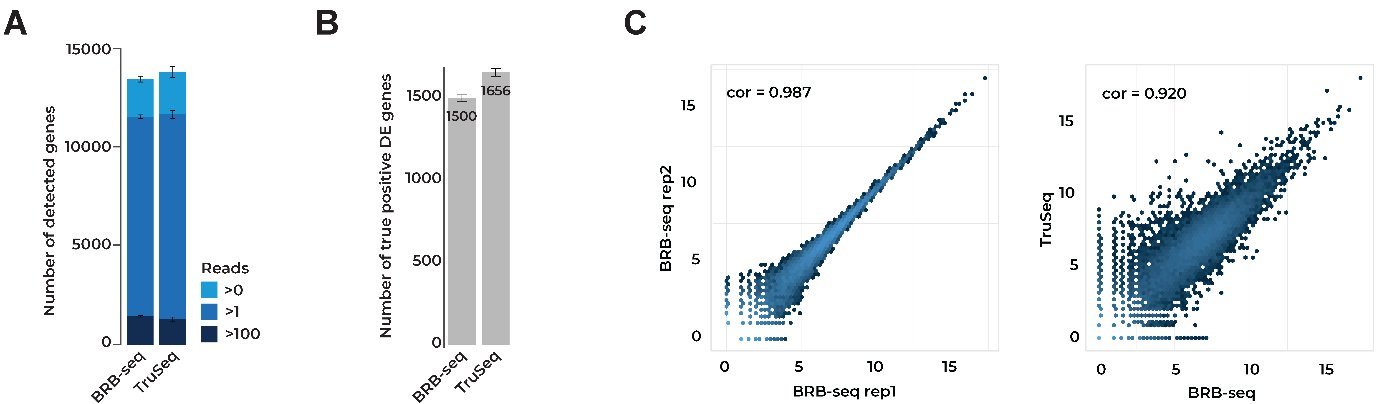
In our original BRB-seq publication, we compared BRB-seq to the ‘gold standard’ Illumina TruSeq Stranded mRNA library preparation method for bulk transcriptomics (Alpern et al., 2019).
At the same sequencing depth, both approaches found similar numbers of expressed genes (Fig. 1A), differentially expressed (DE) genes (Fig. 1B), and a similar correlation between replicates (Fig. 1C).