CUT&Tag (Cleavage Under Targets & Tagmentation) is a breakthrough strategy that combines immunotethering and Tn5-based tagmentation technologies for ultra-sensitive epigenomic mapping1-3. By eliminating the most frustrating parts of ChIP-seq, including chromatin fragmentation, IP, and library prep, CUT&Tag enables streamlined chromatin profiling with less optimization and improved throughput. The high sensitivity of CUT&Tag dramatically reduces background signal, which lowers cell input requirements, sequencing depths, and overall assay costs.
A look inside the CUTANA™ CUT&Tag Kit
EpiCypher is leading the development of CUT&Tag protocols, enzymes, and supporting reagents, and recently launched the CUTANA™ CUT&Tag Kit. This kit is supported by more than two years of research and development and includes multiple controls to help ensure robust performance. In this blog we cover the main features and advantages of the CUTANA CUT&Tag Kit and provide resources to help get you started.
The CUTANA™ CUT&Tag Kit: Everything you need in one box
EpiCypher’s CUTANA™ CUT&Tag Kit delivers a one-stop solution for reliable CUT&Tag profiling making this breakthrough technology widely accessible to the research field. Features include:
- Single-tube workflow with no library prep: The CUTANA CUT&Tag Kit boasts an exclusive Direct-to-PCR workflow2,3, making it the most streamlined CUT&Tag assay on the market (see below).
- Scalable, two-day protocol: The kit contains everything you need to go from cells to sequence-ready libraries in <2 days and uses 8-strip tubes and multi-channel pipettors to support greater throughput.
- Reduced sequencing costs: Only 5-8 million paired-end sequencing reads are needed for robust CUT&Tag profiles vs. 30 million reads for ChIP-seq.
- Expanded sequencing plex: Our indexing strategy allows the entire 48-reaction kit to be pooled in a single run. Combined, the two versions of the kit (14-1102 & 14-1103) generate 96 unique libraries.atures and advantages of the CUTANA CUT&Tag Kit and provide resources to help get you started.
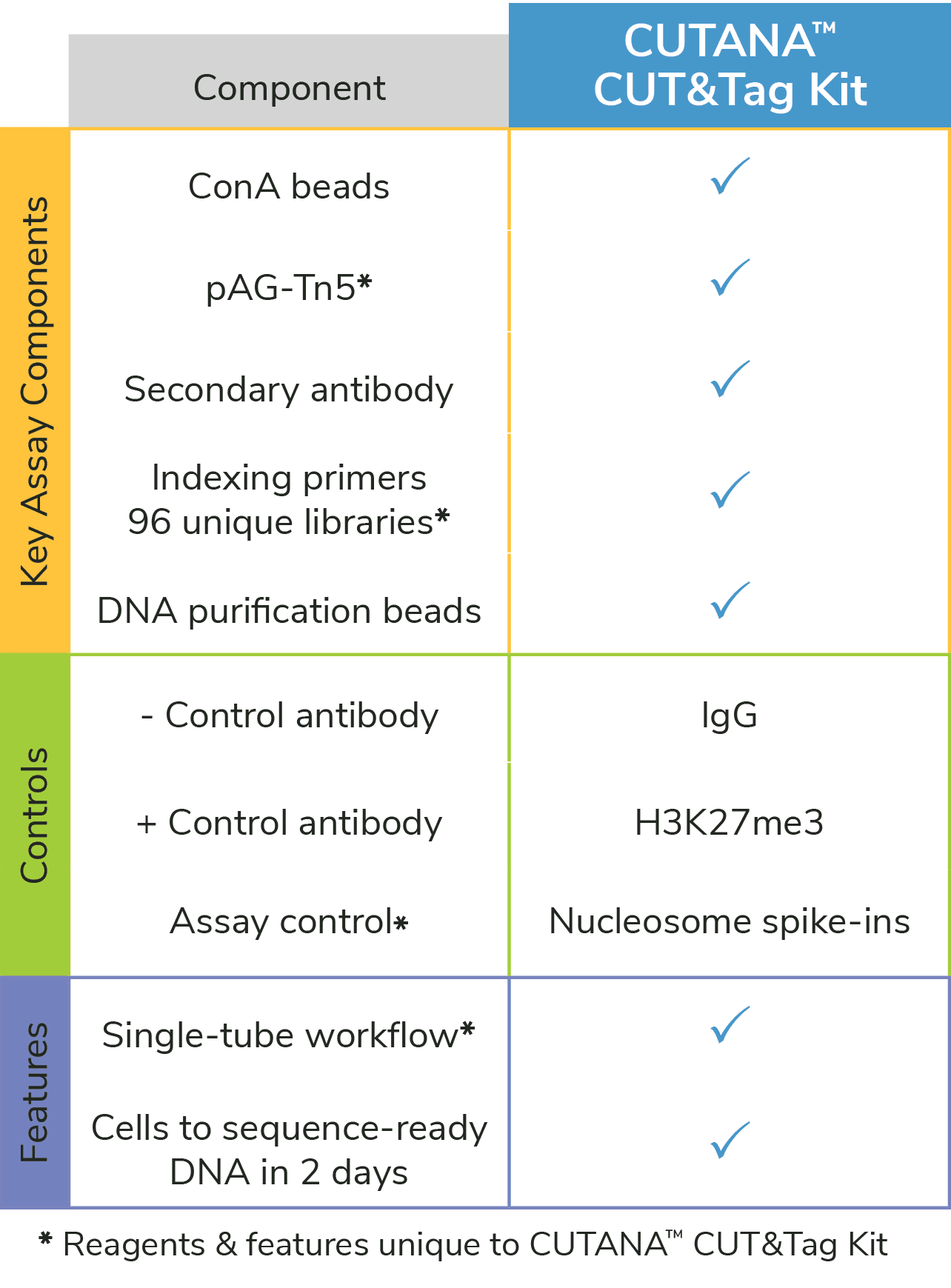
Figure 1: The CUTANA™ CUT&Tag Kit contains essential materials to go from cells to sequence-ready libraries. *Unique features of the EpiCypher CUTANA CUT&Tag Kit.
- Compatible with low inputs: CUTANA CUT&Tag is validated down to 10,000 nuclei, making it compatible with rare/precious cell types (see below) and allowing you to map more targets per sample.
- Assay-specific controls: Validated controls are included for workflow development, monitoring, and troubleshooting (see below). Use these controls in every CUT&Tag experiment for high-quality results.
- Comprehensive manual: Our manual contains all the information you need for successful CUT&Tag experiments, including variations for sample types and detailed optimization guidelines (below).
NOTE: The CUTANA CUT&Tag Kit is most robust for histone post-translational modifications (PTMs). For chromatin-associated proteins we recommend CUT&RUN, which generates robust profiles across diverse target classes and sample types.
Streamlined single-tube CUT&Tag protocol
The CUTANA CUT&Tag Kit utilizes EpiCypher’s exclusive Direct-to-PCR CUT&Tag protocol, allowing users to go from cells to PCR amplified libraries in a single tube2,3. To further increase experimental reproducibility and throughput, the CUTANA CUT&Tag Kit is also optimized for multi-channel pipetting and 8-strip tubes. The entire protocol can be completed in less than two days, and combined with the expanded multiplexing capabilities, allows users to generate more data in less time. This represents a fraction of the effort required for ChIP-seq, which has been notoriously difficult to scale due to high assay costs, laborious workflows, slow processing times, and poor data quality.
Why does the single-tube protocol matter?
In traditional CUT&Tag workflows, phenol-chloroform extraction2, spin columns or DNA-binding magnetic beads1 are used to purify total DNA following antibody-targeted tagmentation. Total DNA is used for indexing PCR, where adapter-ligated DNA is selectively amplified and barcoded, and final sequencing libraries are purified using magnetic beads2.
EpiCypher’s protocol eliminates the time-consuming DNA purification step following tagmentation. Instead, tagmented DNA fragments are directly amplified from the digested nuclear pellet (Figure 2). After indexing PCR, DNA libraries are purified using a bead-based cleanup.
This Direct-to-PCR method streamlines CUT&Tag and mitigates sample loss caused by intermediate DNA purification steps, supporting the use of low nuclei/cell numbers. This strategy has even been leveraged for automated CUT&Tag analysis2,5.
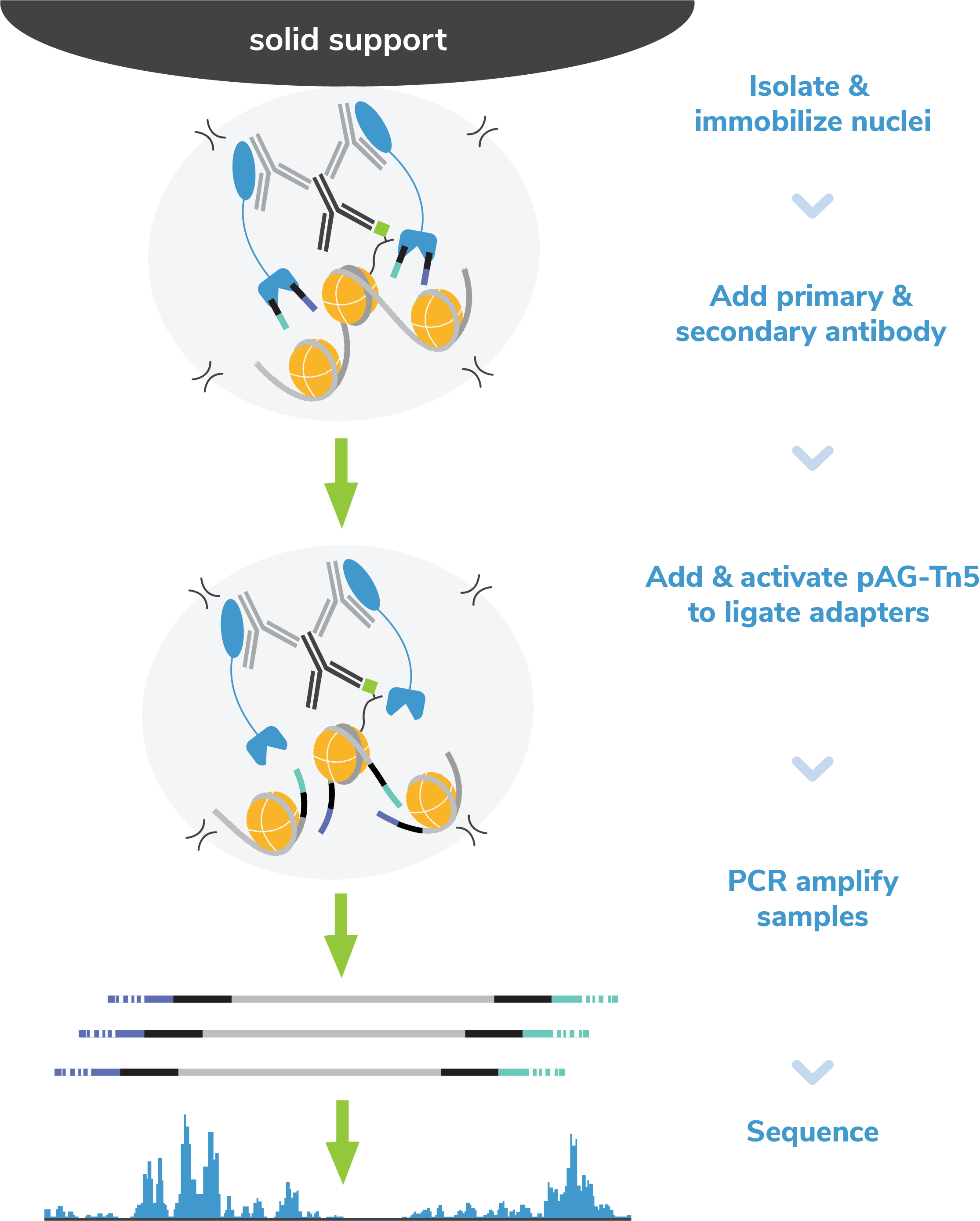
Figure 2: EpiCypher’s exclusive single-tube CUT&Tag protocol streamlines the workflow and reduces sample loss caused by DNA extraction methods, enabling increased assay sensitivity.
Validation for low cell numbers
EpiCypher recognizes the demand for reliable profiling from low cell numbers. In fact, CUT&Tag was specifically developed by the Henikoff lab to enable high-quality, cost-effective chromatin mapping from ultra-low cell inputs1. The use of tagmentation and the Direct-to-PCR protocol eliminates multiple assay steps, reducing the risk of sample loss and making CUTANA CUT&Tag well suited for low sample inputs.
EpiCypher has validated our CUT&Tag Kit for reliable profiling using 100,000 to 10,000 nuclei and has the data to back it up. As shown in Figure 3, EpiCypher’s CUT&Tag kit generates reliable data for histone PTM targets of high (H3K27me3) and low (H3K4me1) abundance. Our kit manual provides detailed guidance and considerations for reducing cell inputs.
*What about inputs below 10,000 nuclei/cells? Although CUT&Tag has been used for single cell or low-input profiling1,3,5-9, these data were generated using microfluidics platforms and/or combinatorial barcoding. With the CUTANA CUT&Tag Kit, most users require at least 10,000 nuclei for reproducible CUT&Tag profiles. Note that success at low inputs is highly dependent on antibody specificity and efficiency, target abundance in your cell type, and the quality of sample preparation.
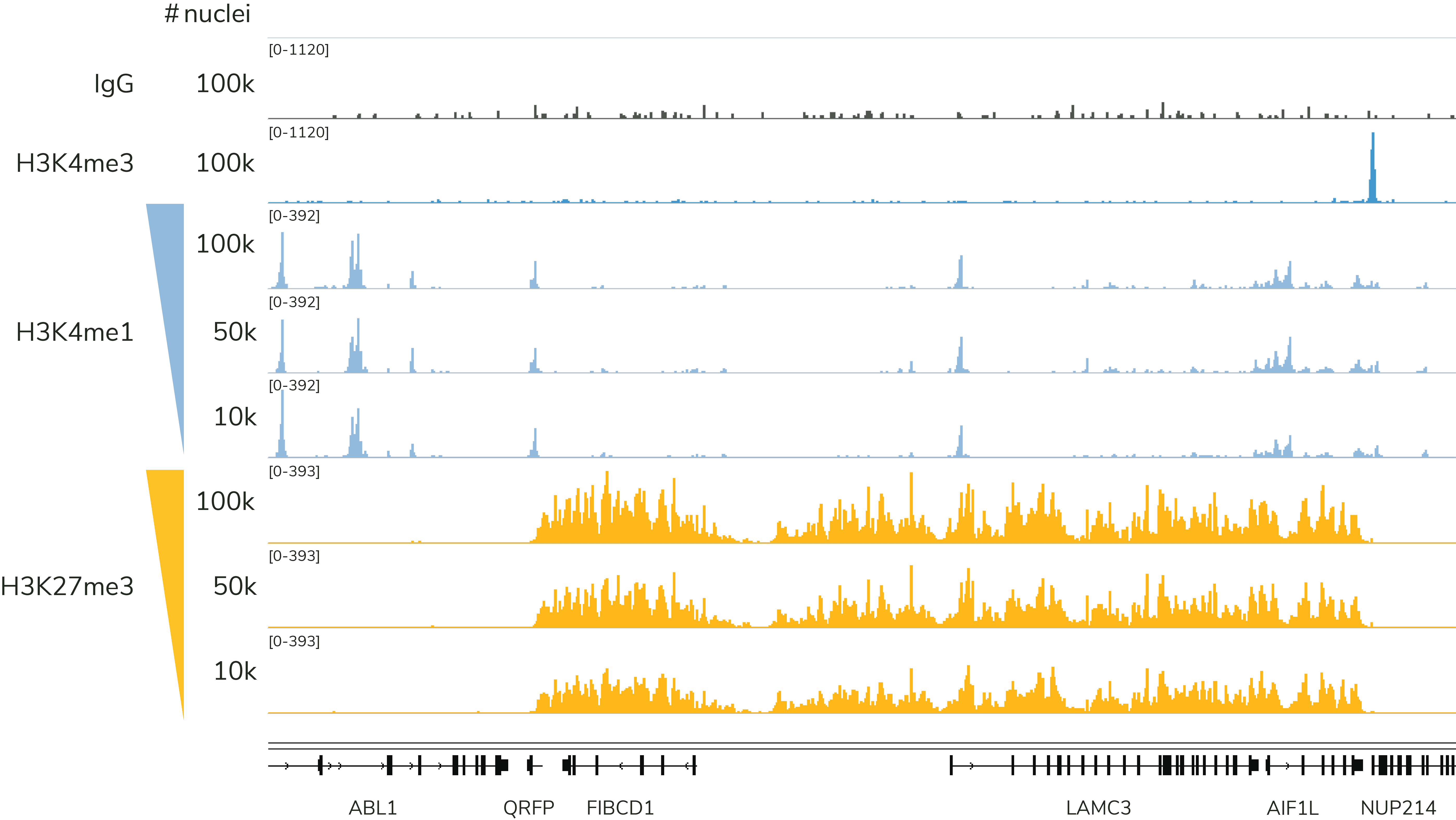
Figure 3: The CUTANA CUT&Tag Kit yields robust, reliable profiles down to 10,000 nuclei. Representative genome browser tracks are shown for H3K4me1 (low abundance target; EpiCypher Cat No. 13-0057) and H3K27me3 (high abundance target; EpiCypher Cat No. 13-0055). CUT&Tag data were generated by EpiCypher using decreasing amounts of K562 nuclei and 5-8 million sequencing reads per reaction.
Reliable CUT&Tag assay controls & success metrics
Controls are essential for all chromatin mapping assays. Due to the differences between CUT&Tag and ChIP-seq approaches, it isn’t always clear what controls are needed. EpiCypher’s CUTANA CUT&Tag Kit and manual include multiple controls to ensure robust experiments (Figures 4 & 5).
Control antibodies and SNAP-CUTANA Spike-in Controls
- H3K27me3 Positive Control Antibody, validated for robust performance in CUT&Tag down to 10,000 nuclei. This antibody is an ideal positive control when developing CUT&Tag workflows for new targets, sample types, or reduced cell numbers. We recommend including the H3K27me3 positive control paired with the K-MetStat Panel (see below) in every experiment – even for previously validated workflows – to monitor assay reliability and troubleshoot any problems that may arise.
- IgG Negative Control Antibody, a key control used to examine background signal and support robust peak calling. Importantly, the Tn5 transposase applied in CUT&Tag naturally prefers accessible chromatin. Although the high-salt buffer used during tagmentation minimizes signal at open chromatin, some background may still be present. The IgG control antibody allows users to detect assay background and incorporate this information for accurate peak calling.
- SNAP-CUTANA™ K-MetStat Panel, a pool of 16 DNA-barcoded nucleosomes (me1, me2, and me3 on H3K4, H3K9, H3K27, H3K36, and H4K20, plus an unmodified control) that is spiked into reactions designated for positive and negative control antibodies (Figure 4). The K-MetStat Panel is the only control that uses purified recombinant nucleosomes, replicating the physiological target of CUT&Tag and providing reliable on- and off-target substrates for control reactions.
When used together, the K-MetStat Panel of spike-ins and the control antibodies provide a powerful tool and form the foundation of EpiCypher’s robust quality control strategy for CUT&Tag experiments. Results are used to validate workflows, confirm control antibody performance, determine assay success, and flag failed reactions. The CUT&Tag Kit Manual details the spike-in workflow, analysis, and how to use the K-MetStat Panel for troubleshooting experiments. By identifying poor samples or reactions and using the spike-in results to guide troubleshooting, researchers can be confident in their experimental results.
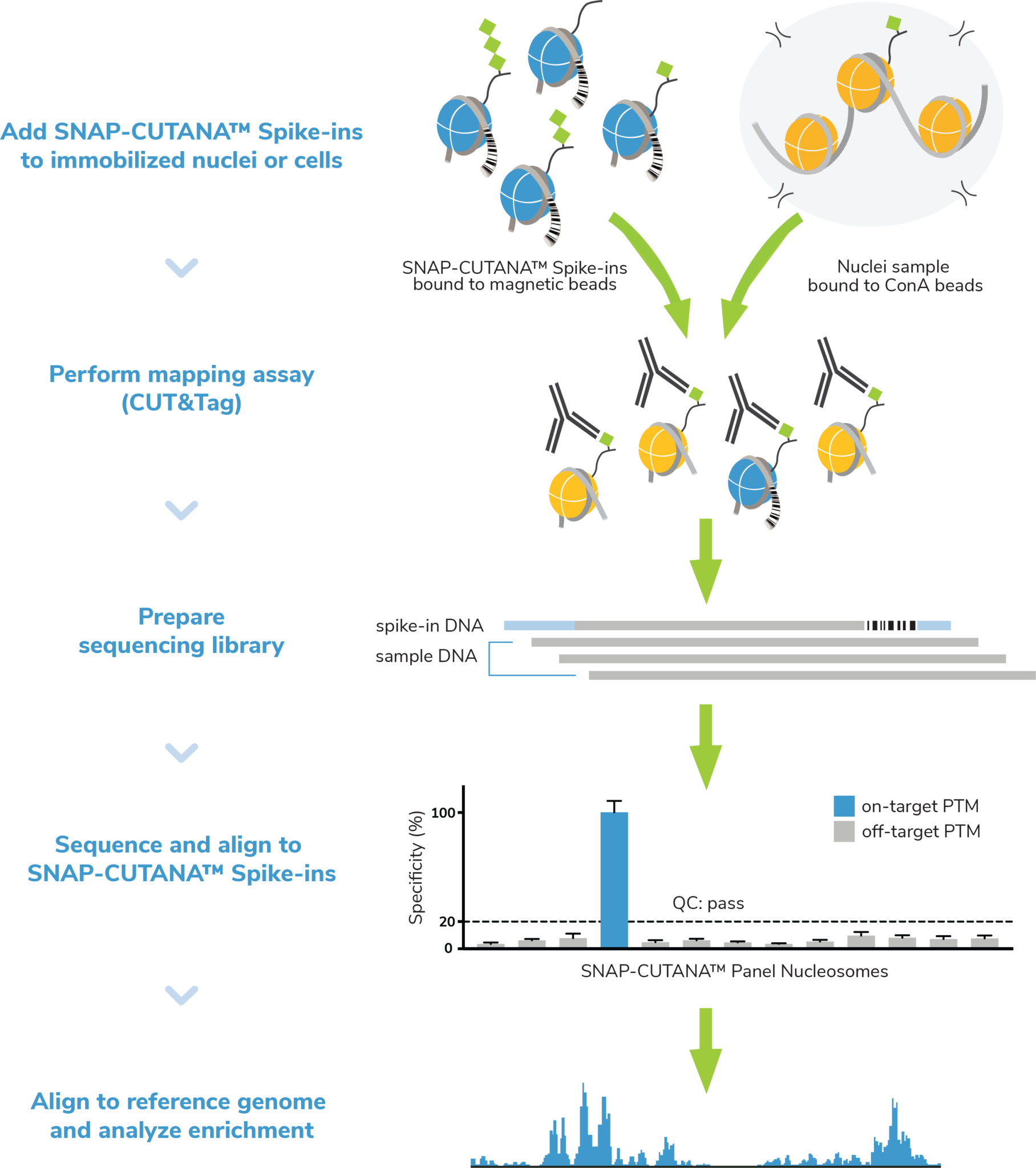
Figure 4: The SNAP-CUTANA K-MetStat Panel of nucleosome spike-ins and control antibodies are an essential tool for CUT&Tag workflow validation and determination of experimental success. Here, nuclei are coupled to ConA beads, spiked with the K-MetStat Panel, and treated with an antibody (e.g. H3K27me3), which binds its target histone PTM in both sample chromatin AND in the K-MetStat Panel. Targeted tagmentation and indexing PCR generate sequencing libraries containing fragments from sample chromatin AND the K-MetStat Panel. Sequencing data are aligned to PTM-specific DNA barcodes, revealing specific recovery of the on-target spike-in with high signal over background, thus validating the entire CUT&Tag workflow. Sequencing data are then aligned to the species genome and subjected to additional analysis (peak calling, etc.).
Quality Control Checks
Our manual contains quality control checks designed to flag problems at critical steps in CUT&Tag experiments. The earlier problems can be identified, the easier it is to determine the cause and select the appropriate troubleshooting steps. Each of our quality control checks include guidance on troubleshooting to help users improve workflows. We outline the most important steps below and in Figure 5.
- Sample prep and binding to ConA beads: We use a simple Trypan Blue staining method to confirm starting cell viability/morphology, nuclear integrity (i.e. no lysis), and binding to ConA beads. You can’t expect to generate high quality results if starting with poor quality samples. In fact, if any one of these steps goes awry, the entire CUT&Tag experiment is at risk for failure.
- Success metrics before sequencing: The best indicator of success before sequencing is enrichment of mononucleosome-sized fragments (~300 bp including sequencing adapters) in PCR-amplified libraries, as shown by Agilent® Bioanalyzer or TapeStation fragment distribution analysis. As noted in our manual, library yields are NOT a reliable measure of assay success, as yields vary widely based on cell type, quality of sample prep, antibody specificity/efficiency, and target abundance. As long as you generate sufficient library for Illumina next-generation sequencing (≥2 ng/µL), you are good to go!
- Success metrics after sequencing: We rely on the control reactions (see above) to examine CUT&Tag experiments post-sequencing (Figure 5), outlined in further detail in the kit manual. For experimental targets, EpiCypher suggests aiming for 5-8 million paired-end sequencing reads. Data should have high signal, low background, and show expected enrichment and/or peak patterns.
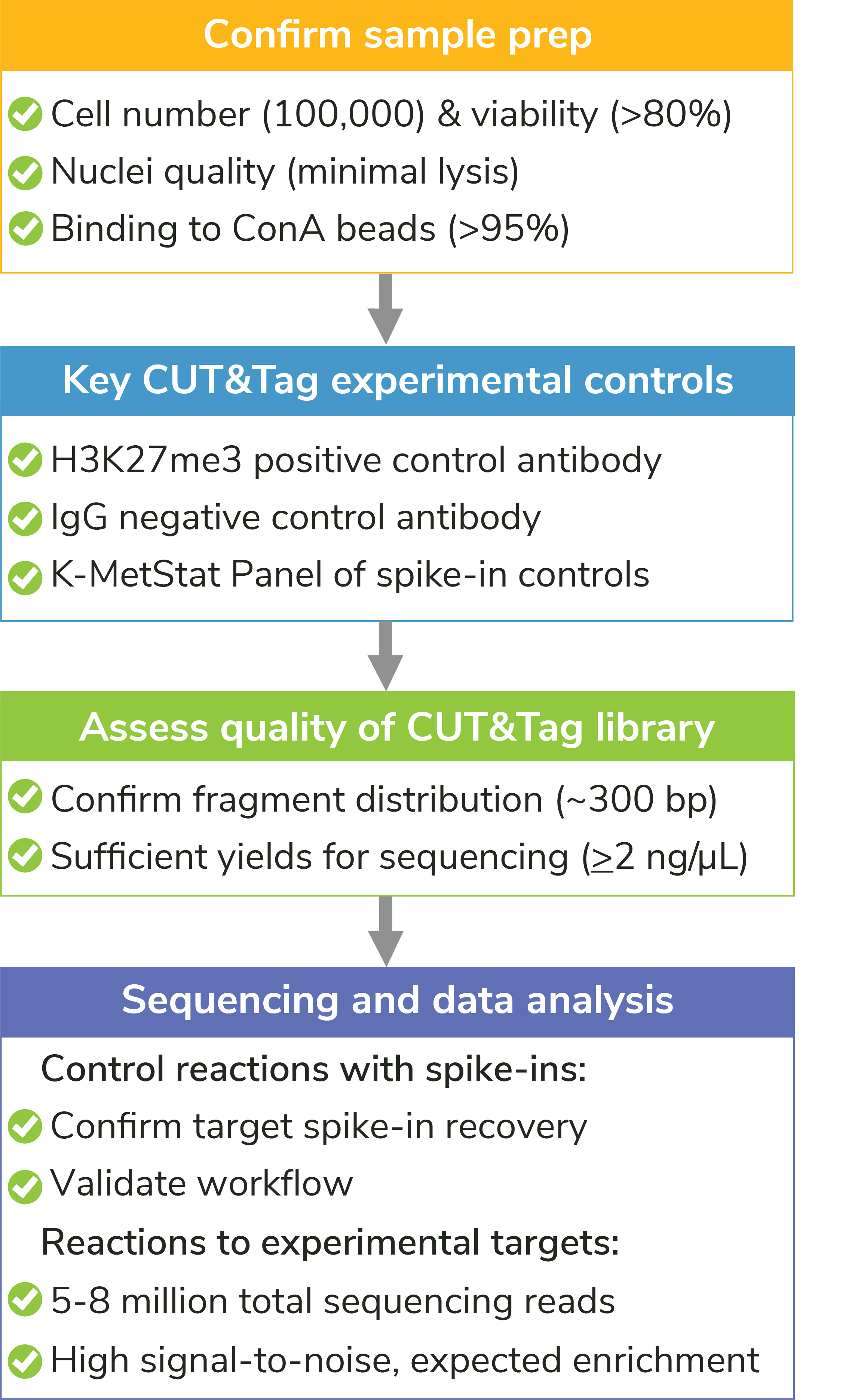
Figure 5: Quality control checks included in the CUTANA CUT&Tag Kit protocol help ensure experimental success, flag potential problems, and guide troubleshooting experiments. For detailed information about each check, see the CUTANA CUT&Tag Kit manual.
Comprehensive manual with protocol variations, key method notes, troubleshooting guidance, and more
Another goal of the CUTANA CUT&Tag Kit is to make CUT&Tag more user-friendly. The kit manual incorporates everything our scientists have learned through developing CUT&Tag protocols and kits, making it an essential guidebook for exploring CUT&Tag applications.
Protocol variations:
Our Direct-to-PCR CUT&Tag protocol was originally developed using nuclei from human K562 cells. However, we have included protocol alterations for:
- Whole cells: Nuclei are preferred but cells can also be used.
- Cryopreserved cells and nuclei: We provide best practices for freezing and thawing samples.
- Crosslinking: Light cross-linking conditions can be used (Figure 6) – but may reduce yields.
- Adherent cells: Our simple trypsin-based procedure can be used to collect adherent cells.
- Tissues: It is essential to generate a single cell suspension from tissues; references are provided.
- Immune cells: ConA beads can activate some immune cell types – we recommend using nuclei.
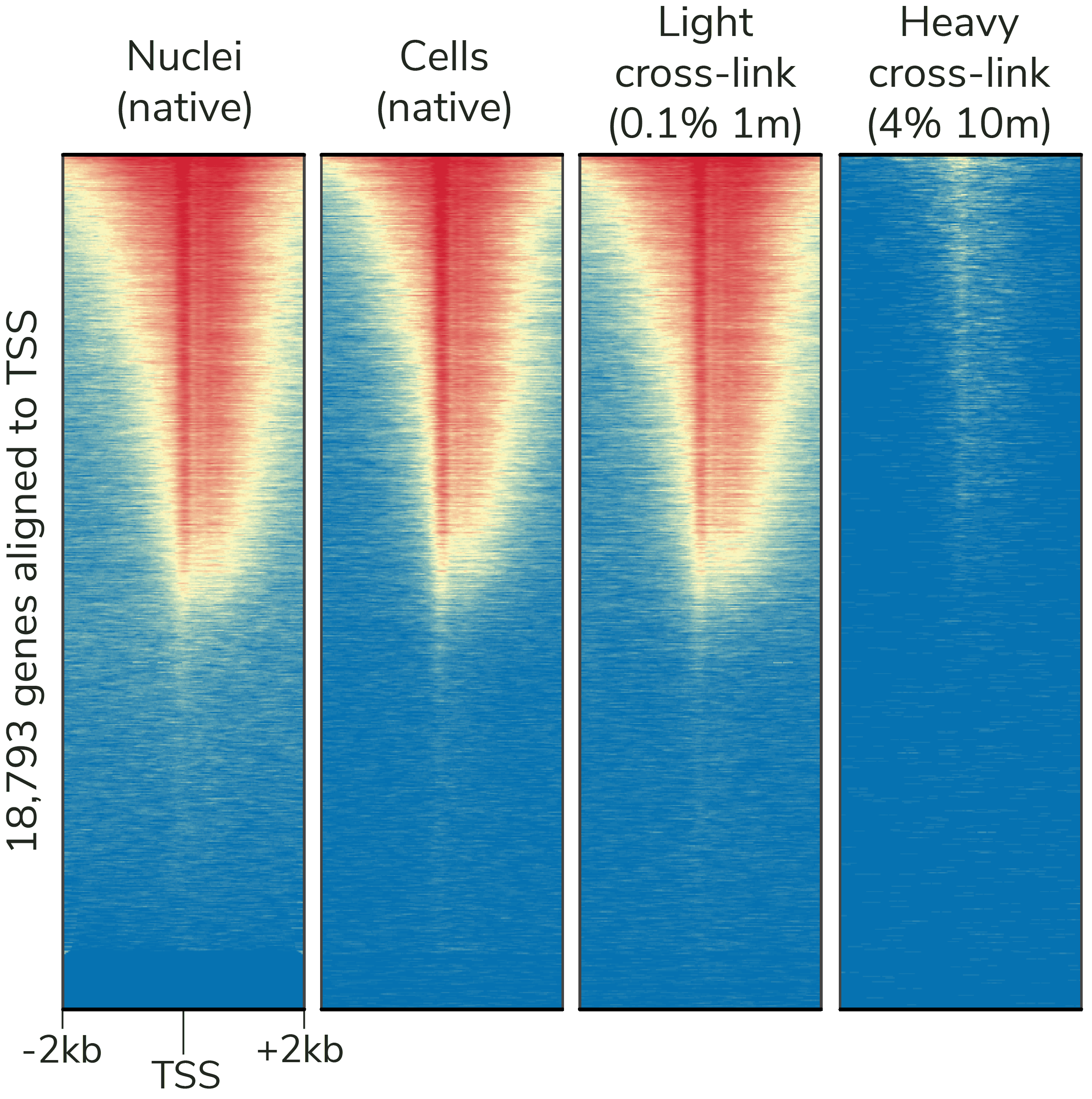
Figure 6: CUTANA CUT&Tag assays generate robust results with native and lightly cross-linked samples, while ChIP-seq cross-linking conditions (far right) reduce DNA yields. CUT&Tag data were generated using an H3K4me3 antibody (EpiCypher Cat No. 13-0041) and 100,000 K562 cells or nuclei per reaction. Heatmaps show signal intensity around the transcription start site (TSS) of 18,793 genes, from blue (low signal) to red (high signal). Gene rows are aligned across conditions.
Chromatin accessibility profiling with CUT&Tag:
Mapping accessible chromatin regions provides valuable context for epigenetics research, especially when paired with histone PTM profiles marking distinct genomic features. ATAC-seq – the most popular assay – suffers from high levels of mitochondrial DNA contamination, leading to increased assay background and high sequencing costs (>50 million reads per reaction).
The CUTANA™ CUT&Tag Kit Manual includes the CUTAC (Cleavage Under Target Accessible Chromatin) protocol, an EpiCypher-exclusive approach that leverages pAG-Tn5 for high-resolution, low-cost mapping of accessible chromatin. CUTAC only requires 5-8 million sequencing reads, has low mitochondrial read contamination, and shows exquisite signal over background, providing an attractive alternative to ATAC-seq profiling3. The workflow changes are minimal, allowing you to perform standard CUT&Tag assays and CUTAC assays in parallel, if desired.
Key protocol notes:
Helpful tips from our genomics scientists are provided at sample prep, tagmentation, indexing PCR, and library analysis steps to support successful CUT&Tag experiments.
For example, proper handling of nuclei immobilized on Concanavalin A (ConA) beads is crucial to experimental success. Clumping of ConA beads remains one of the most common problems in CUT&Tag assays. Although some ConA bead clumping is normal, excessive bead aggregation leads to bead dry out, sample loss, and low yield. We have identified multiple solutions to this problem. Suggestion include:
- Avoid cell/nuclear lysis, which increases bead clumping! Take care when resuspending cells and ConA beads, especially for delicate primary cells or cross-linked samples.
- Add digitonin to buffers as instructed, which prevents ConA beads from forming a film on the side of tubes and drying out.
- Use a nutator for all incubations, which gentle agitates tubes side to side. End-over-end rotation inevitably causes ConA beads to stick to the lids of tubes, resulting in sample loss.
- Thoroughly resuspend ConA beads before pAG-Tn5 incubation, tagmentation, and indexing PCR. If possible, use low-retention filter pipette tips, which are designed for viscous solutions.
- Vortex (do NOT pipette) ConA beads at certain steps where the solution is sticky – especially after tagmentation. These steps are clearly indicated in the manual.
Our Tech Support Team has additional tips and tricks for specific use cases, so if these suggestions don’t help, please reach out to technical@stratech.co.uk – we are confident we can help!
Assay optimization and troubleshooting
In addition to key protocol notes, the manual contains EpiCypher’s recommendations for workflow development (Figure 7), troubleshooting poor results (Appendix 1), antibody validation (FAQs), and sequencing analysis (FAQs). We have also reviewed our suggestions for antibody validation in this blog.
NOTE that all these steps rely heavily on the control antibodies, spike-ins, and quality control checks included in the kit! (See above)
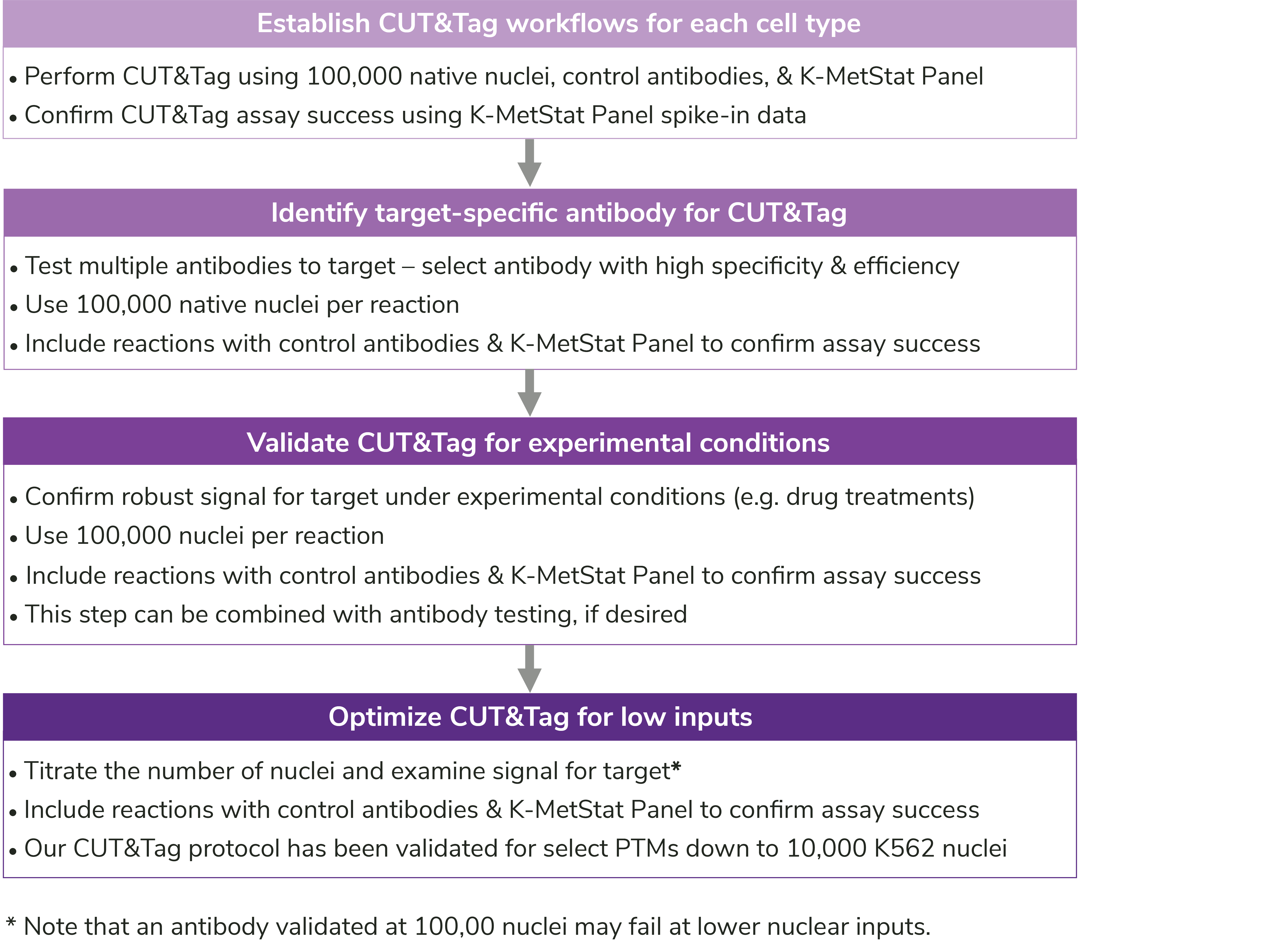
Figure 7: User-specific optimization steps outlined in the CUTANA CUT&Tag Kit Manual.
Ready to get started?
CUT&Tag provides dramatic breakthroughs in chromatin mapping technology. Even when compared to low-input ChIP-seq assays, CUT&Tag still provides multiple benefits:
- Fast: CUT&Tag goes from cell to sequencing libraries in less than two days vs. a week or more using ChIP-seq.
- Streamlined: EpiCypher’s one-tube protocol is designed for straightforward assay setup and rapid sample processing. ChIP-seq was not built with any of these capabilities in mind.
- Reliable, less optimization: CUT&Tag eliminates cell lysis, chromatin fragmentation, IP & harsh wash buffers, and library prep – skipping the most time-consuming and variable parts of ChIP-seq.
- High quality data at low sequencing costs: CUT&Tag only requires 5-8 sequencing reads for high resolution profiles. ChIP-seq typically requires 30 million reads – and still generates poor data!
EpiCypher customers have been using CUTANA CUT&Tag assays to study T cell activation & exhaustion10, SARS-CoV-2 infection11, pancreatic beta cell development12, and much more!
References
- Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10, 1930 (2019). PubMed PMID: 31036827.
- Kaya-Okur HS et al. Efficient low-cost chromatin profiling with CUT&Tag. Nat Protoc 15, 3264-83 (2020). PubMed PMID: 32913232.
- Henikoff S et al. Efficient chromatin accessibility mapping in situ by nucleosome-tethered tagmentation. Elife 9, (2020). PubMed PMID: 33191916.
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10, 669-80 (2009). PubMed PMID: 19736561.
- Janssens DH et al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia. Nat Genet 53, 1586-96 (2021). PubMed PMID: 34663924.
- Janssens DH et al. CUT&Tag2for1: a modified method for simultaneous profiling of the accessible and silenced regulome in single cells. Genome Biol 23, 81 (2022). PubMed PMID: 35300717.
- Deng Y et al. Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science 375, 681-6 (2022). PubMed PMID: 35143307.
- Wu SJ et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat Biotechnol (2021). PubMed PMID: 33846646.
- Bartosovic M et al. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat Biotechnol (2021). PubMed PMID: 33846645.
- Battistello E et al. Stepwise activities of mSWI/SNF family chromatin remodeling complexes direct T cell activation and exhaustion. Mol Cell (2023). PubMed PMID: 36944333.
- Wei J et al. Pharmacological disruption of mSWI/SNF complex activity restricts SARS-CoV-2 infection. Nat Genet 55, 471-83 (2023). PubMed PMID: 36894709.
- Bohuslavova R et al. ISL1 controls pancreatic alpha cell fate and beta cell maturation. Cell Biosci 13, 53 (2023). PubMed PMID: 36899442.