CUT&RUN Support Center
The basics of CUT&RUN
Cleavage Under Targets & Release Using Nuclease (CUT&RUN) is a groundbreaking approach for ultra-sensitive mapping of chromatin targets developed by the group of Dr. Steven Henikoff (see the paper). It builds on immunotethering technology (chromatin immunocleavage or ChIC) developed by Dr. Ulrich Laemmli (see this paper), wherein a fusion of Protein A to Micrococcal Nuclease (pA-MNase) is recruited to selectively cleave antibody-bound chromatin in intact cells or nuclei.
In CUTANA CUT&RUN, cells or nuclei are immobilized to a solid support and Protein A/Protein G-MNase (pAG-MNase) is used to selectively cleave antibody-labeled chromatin in intact cells (see Figure 1 for workflow overview). The clipped fragments diffuse into solution, where they can be separated from cells, purified, and analyzed by next-generation sequencing. This workflow results in high-quality, genome-wide profiles of histone post-translational modifications (PTMs) and chromatin-associated proteins (e.g. transcription factors).
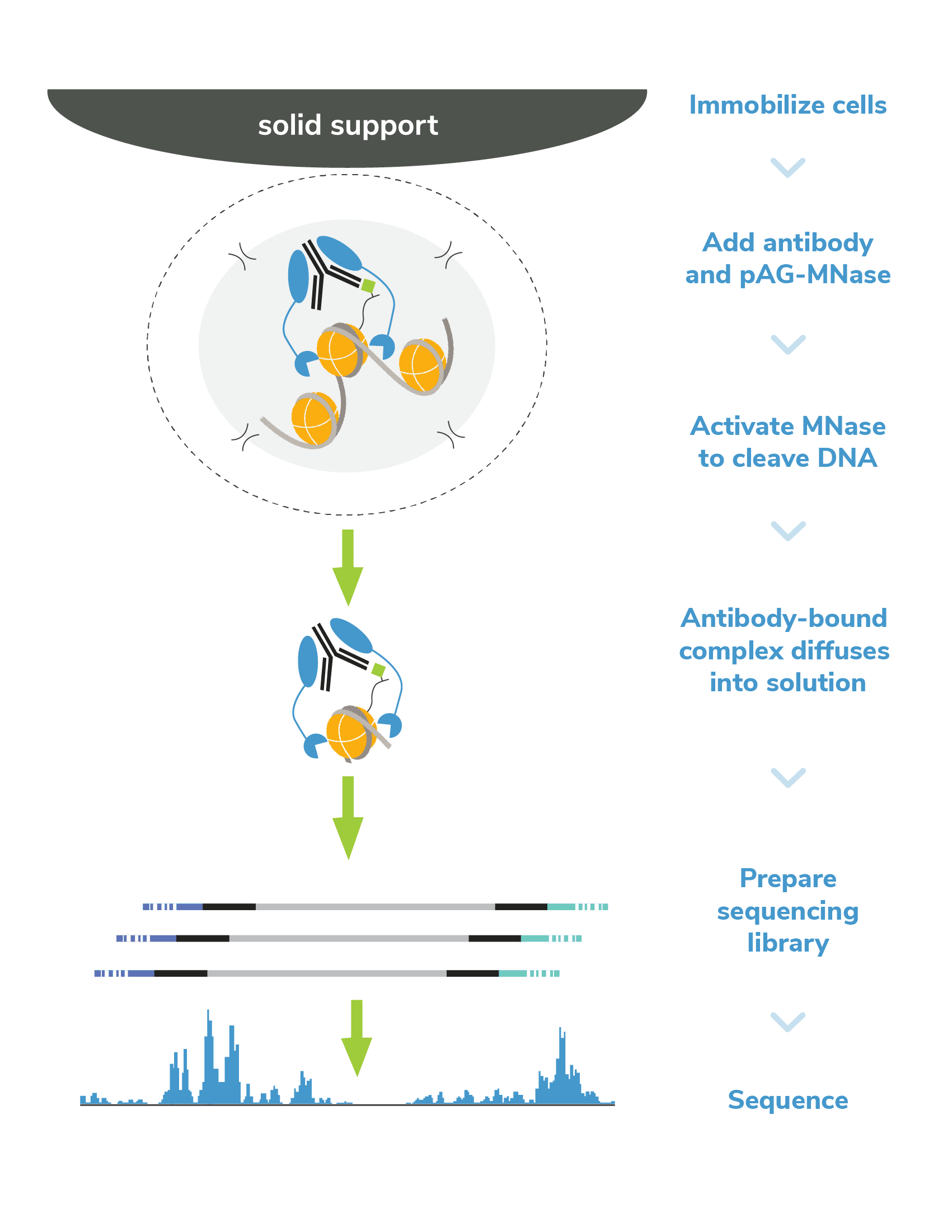
Figure 1. Overview of CUTANA CUT&RUN workflow.
ChIP-seq and CUT&RUN are both used to map histone PTMs and protein-DNA interactions, but they are not equivalent strategies. ChIP-seq, or chromatin immunoprecipitation sequencing, uses as antibody to enrich or “pull-down” targets from a large pool of fragmented chromatin. Generating the chromatin pool requires cross-linking, cell lysis, and chromatin fragmentation, each of which must be carefully optimized and controlled to reduce artifacts and background signal. Following fragmentation, bead-coupled antibodies are used to pull down target fragments. Cross-links are reversed and DNA is purified for sequencing analysis. Chromatin immunoprecipitation (the IP step) is inherently noisy. In addition to problems with antibody performance, the magnetic beads may pull down off-target fragments along with the chromatin containing the target. IP also requires highly stringent washes, which can reduce yields and lower signal-to-noise.
CUT&RUN revolutionized the study of chromatin regulation by enabling targeted release of genomic fragments into solution. The streamlined workflow does not require sonication or IP steps, resulting in dramatically reduce background and improved on-target recovery. The result is an assay amenable to greater experimental throughput, allowing deeper and more rapid investigations to uncover epigenetic biology.
Specifically, compared to ChIP-seq, CUT&RUN features:
- Lower cell requirements. CUTANA™ CUT&RUN assays generate high-resolution profiles using as few as 5,000 cells, and lower cell numbers have been reported in the literature (PubMed PMIDs: 30955888, 33911257).
- Improved signal-to-noise. Separation of target-bound chromatin from bulk material means lower background and robust signal – even when using fewer cells.
- Reduced cost. High signal-to-noise enables confident peak calling with ~10-fold fewer sequencing reads and less primary antibody.
- Less optimization. The entire assay is performed in intact cells (or nuclei) immobilized on a solid support. These advances eliminate the most technically challenging steps of ChIP-seq, including cell lysis, chromatin fragmentation, and IP.
- Faster. CUT&RUN provides a user-friendly, streamlined protocol that allows scientists to go from cells to sequencing data in less than four days (vs. weeks for ChIP-seq). CUT&RUN can also be automated for greater throughput.
CUT&RUN consists of just a few basic steps!
Step 1: Isolate cells & immobilize to concanavalin A (ConA) beads
Cells (or nuclei) are bound to magnetic beads coated with Concanavalin A (ConA), a lectin that binds to cell surface proteins. This supports high-throughput formatting and simplifies the separation of cells from clipped chromatin fragments in later steps.
It is important to avoid bead dry out and clumping, as this results in sample loss and reduces yields. Important quality control checks to confirm cell integrity and ConA bead binding can be found here. Additionally, some sample types may require modifications; see Sample Prep for more information.
Step 2: Permeabilize cells
Immobilized cells are treated with a buffer containing Digitonin, a nonionic detergent that permeabilizes cell membranes at low concentrations. Permeabilization is crucial for antibody and pAG-MNase binding, and allows MNase-digested DNA to diffuse into solution in later steps. Digitonin conditions must be optimized for each cell type to avoid cell lysis and/or incomplete permeabilization. For information about optimization of this step click here.
Step 3: Incubation with target-specific antibody
An antibody to the target of interest is added to the reaction and incubated overnight at 4˚C. We suggest including negative control (e.g. IgG) and positive control (e.g. H3K4me3) reactions in every experiment.
The specificity and binding efficiency of antibodies is crucial for successful CUT&RUN. In fact, the background of this assay is so low that an antibody with poor efficiency will not generate high enough yields for PCR and sequencing. In contrast, a nonspecific antibody may provide decent yields, but will lead to incorrect biological interpretations. See additional notes on antibody selection and assay controls to learn more.
Step 4: pAG-MNase binding
The following day, bead-bound cells are washed to remove unbound and/or non-specifically bound antibody. pAG-MNase is added to the reaction in the absence of calcium (Ca2+) to prevent premature activation of MNase. The immunoglobulin binding properities of pAG act to “tether” MNase to antibody-bound chromatin. Following pAG-MNase incubation, the cell/bead mixture is washed several times to remove excess pAG-MNase, which helps to prevent non-specific cleavage.
Step 5: pAG-MNase activation
Ca2+ is added to the reaction to activate MNase, which cleaves DNA proximal to where the antibody is bound. Cleaved chromatin fragments diffuse into the supernatant, while remaining bulk chromatin remains inside the bead-immobilized cells.
Because MNase is a processive enzyme, the reaction must be quenched to prevent over-digestion of released DNA. Following pAG-MNase incubation, a Stop Buffer containing EDTA and EGTA is added to chelate free calcium ions and halt enzymatic activity. Reactions are briefly heated to degrade RNA and release any remaining chromatin fragments into solution.
Step 6: DNA purification
Isolation of CUT&RUN enriched DNA is straightforward because the cells remain bound to magnetic ConA beads. Bead-coupled cells containing bulk chromatin are magnetically separated from the clipped target DNA, which remains in solution. Target DNA is purified and quantified with a fluorometric assay .
DNA yields should NOT be used as an indicator of CUT&RUN success. Instead aim for ~5 ng DNA, which will allow robust CUT&RUN library prep. It is NOT recommended to analyze raw CUT&RUN DNA on the Bioanalyzer/TapeStation, as yields are often below the limit of detection for these methods.
EpiCypher also checks that the yields from positive controls and experimental targets are greater than the IgG negative control reaction, even if only slightly higher. After confirming DNA quality according to our outlined metrics, proceed to library prep.
Step 7: CUT&RUN library prep
Purified CUT&RUN DNA is repaired, ligated to sequencing adapters, and PCR-amplified to generate sequencing libraries. PCR is performed using parameters optimized for low CUT&RUN yields and small fragment sizes, and barcoded primers are used to enable multiplexed sequencing. EpiCypher’s Library Prep Kit is specifically optimized to further streamline your workflow.
Prior to sequencing, the best method to confirm CUT&RUN success is fragment size distribution analysis of purified libraries. The fragment size distribution and concentration of CUT&RUN libraries is confirmed using capillary electrophoresis (e.g. Agilent Bioanalyzer or TapeStation). Because MNase digests fragments to nucleosome-level resolution, the average peak size is typically ~300 bp (~170 bp fragmented DNA + adapters). For more further reading on assuring sequencing library quality, see this article.
Step 8: Illumina® next-generation sequencing
Libraries are pooled at equimolar ratios and loaded onto the desired platform for sequencing. Only 3-8 million reads per sample are required for robust signal over background (vs. >20 million for ChIP-seq), allowing users to multiplex 10s-100s of samples in a single run.
I. Sample Preparation
Number of Cells. We recommend starting with 500,000 native (unfixed) cells, particularly when mapping new targets or using new cell types.
- Notes on cell types: EpiCypher has developed several protocol variations to ensure reliable CUT&RUN data from diverse biological samples. For recommendations on alternate sample prep protocols (e.g. using adherent cells, tissues, immune cells, nuclei, and frozen cells/nuclei), see this page.
- Notes on using tissues: CUTANA CUT&RUN Kits and protocols are compatible with tissues. The primary requirement when using tissues is to process samples into a monodispersion of cells or nuclei. See this article for more information.
- Notes on low inputs: Following initial validation of workflows using 500,000 cells and control antibodies, cell numbers can be reduced following the recommendations described here.
Replicates. CUT&RUN is very robust and reliable – two biological replicates (e.g. same cell type harvested from two mutant mice) per target are usually sufficient.
Cross-linking. CUT&RUN is a native technique, meaning that it performs best on unfixed cells (or nuclei). This is a major advantage compared to ChIP, which typically requires heavy cross-linking to stabilize target associations with DNA. Cross-linking and chromatin fragmentation are major contributors to high background, low yields, artifacts, and/or data variability in ChIP. These steps aren’t required for CUT&RUN, which streamlines the assay, maximizes on-target DNA recovery, and allows for reduced cell numbers.
However, there are some instances where light cross-linking can be useful in CUT&RUN, by localizing and stabilizing potentially labile PTMs (e.g. histone acetylation) and acetyl-binding proteins (e.g. bromodomains) or when performing time-course or drug treatment assays. Note that we always recommend trying native conditions first, or at least in parallel with cross-linked samples. See this blog and our cross-linking protocol for detailed information.
Optimization of Cell Permeabilization. This is a key step of the CUT&RUN protocol, as the cell membrane must be pervious to antibodies and pAG-MNase, yet intact enough to prevent cell lysis. EpiCypher’s standard CUT&RUN condition for whole cells is 0.01% Digitonin. This may not be sufficient dependeing on cell type (e.g. fibroblast or macrophages). For optimizing Digitonin conditions, use the full step-by-step procedure as outlined here. If you are still experiencing permeabilization issues, try extracting nuclei from your cells.
II. Assay Controls
All CUT&RUN experiments should include appropriate controls, provided in the CUTANA™ CUT&RUN Kit, to evaluate assay success and individual reaction performance.
Quality Control Checks. EpiCypher has incorporated multiple quality control check points to help ensure assay success. For instance, we have outlined a simple Trypan Blue staining protocol to confirm sample binding to ConA beads prior to antibody addition – a critical step of CUT&RUN. A full list of quality control checks, both before AND after sequencing, can be found here.
Spike-in controls. Spike-in controls are essential for all genomics assays. The CUTANA CUT&RUN Kit includes includes E. coli spike-in DNA, which can be added to all reactions as a control for library prep and to aid in sequencing normalization.
For reactions targeting histone PTMs, EpiCypher offers SNAP-CUTANA™ Spike-in Controls. SNAP-CUTANA Spike-in Controls are panels of highly pure nucleosomes, each containing a defined histone PTM and accompanying PTM-specific DNA barcode. The nucleosomes come pre-bound to magnetic beads for simple one-step addition to CUT&RUN workflows, allowing users to examine antibody specificity, signal over background, and assay variability. Panels are currently available for histone lysine methylation PTMs (K-MetStat Panel) and lysine acetylation and extended acyl states are coming soon. Check to make sure that your target is included in the panel before adding to reactions; see our section on SNAP-CUTANA Spike-ins to learn more about how to leverage SNAP-CUTANA Spike-ins for your workflow.
Positive and Negative Control Reactions. Reactions using negative control (IgG) and positive control (H3K4me3) antibodies should be included in every experiment to validate CUT&RUN workflows. EpiCypher also recommends adding the SNAP-CUTANA™ K-MetStat Panel of spike-in controls to these positive and negative control reactions, to provide a direct readout of assay success and to guide troubleshooting experiments. For more information about the SNAP-CUTANA K-MetStat Panel and how it can be used to guide troubleshooting, see this article.
III. Antibody Selection
Select a target-specific antibody. As with ChIP-seq, a quality antibody is essential for generating robust and reliable genomic profiles. Key considerations include:
- Performance in other assays, such as ChIP, does NOT guarantee success in CUT&RUN! The antibody should demonstrate high-specificity (low cross-reactivity) and high efficiency (comprehensive target recognition) in CUT&RUN.
- EpiCypher offers CUTANA™ CUT&RUN antibodies against histone PTMs and multiple target classes of chromatin-associated proteins. These antibodies have been extensively screened for robust performance in CUT&RUN assays.
- When analyzing a new target in the absence of validated antibodies, we recommend sourcing 3-5 antibodies from various reputable vendors that bind unique epitopes. Test these antibodies in parallel CUT&RUN assays with cells known to express the target protein. Then, select the best antibody based on overall yield, signal over background, and peak structure/enrichment.
- Need more information? We cover principles for antibody selection extensively here.
IV. DNA Purification and Analysis
DNA purification. Yields from CUT&RUN are much lower compared to ChIP, and DNA fragments can also be much smaller, particularly for transcription factors. In these cases, it is important to use a DNA purification protocol that is optimized for low DNA concentrations and small fragment sizes. EpiCypher’s CUTANA CUT&RUN Kit includes DNA purification protocols that are specifically designed to capture small fragments and maximize DNA concentration for subsequent library prep.
- Note: We also offer these DNA purification reagents in a separate CUTANA™ DNA Purification Kit!
Library prep. Library prep is a standard part of genomics assays and there are numerous kits and multiplexing strategies available. However, the low yields and small fragments from CUT&RUN make it difficult to utilize existing library prep kits. For instance, it isn’t clear if adapter concentrations should be adjusted for low DNA inputs, or the optimal SPRI bead ratio to use for enriching small CUT&RUN fragments while avoiding adapter-dimer contaminants. To address these concerns, EpiCypher has launched an all-inclusive Library Prep Kit specifically optimized for CUTANA CUT&RUN assays. You can read more about library kit development in this blog post.
Sequencing depth. The number of sequencing reads depends on several factors, including the number of cells, target abundance, and antibody quality. For most targets, 3-8 million paired-end reads are sufficient and will allow you to multiplex more samples per run. If you have further questions about sequencing depths or CUT&RUN library prep, see this article.
To learn more about chromatin structure and its regulation of gene expression, watch below.
Want to learn more about the benefits of CUT&RUN over ChIP-seq?
Trying to understand the difference between CUT&RUN and it’s sister technology CUT&Tag?
To better understand the general steps of chromatin mapping assays, data analysis, and how they compare across ChIP-seq, CUT&RUN, and CUT&Tag:
To learn more about sequencing data analysis:
Protocol video walk-through
Kits
Spike-in Controls
Enzyme and Accessory Products
Antibodies
Negative control
Histone PTMs
Chromatin-associated proteins
Protocols
CUT&RUN and accessory protocols
In this article, find all the steps to take you from cells to library prep-ready DNA using the CUTANA™ CUT&RUN Kit. For library prep and sequencing protocols, see this article.
CUT&RUN Protocol: Day 1
Section I: CUT&RUN buffer prep (~30 min)
Important Notes on Buffer Prep
- Buffers are prepared FRESH on Day 1 of every CUT&RUN experiment.
- 0.01% Digitonin is optimal for permeabilizing K562, MCF7, and A549 cells and is recommended for reactions using nuclei. For other cell types, Digitonin conditions MUST be optimized for efficient cell permeabilization. See this article for instructions.
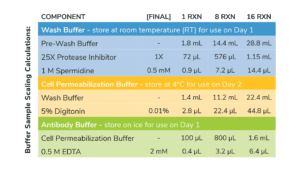
- Volumes in Table 1 are per CUT&RUN reaction and include 20% excess to account for pipetting errors. You do NOT need to add additional volume.
- Gather kit reagents stored at 4˚C and -20˚C needed for Day 1: ConA beads, Bead Activation Buffer, Pre-Wash Buffer, Digitonin, Spermidine, H3K4me3 and IgG control antibodies, K-MetStat Panel. Place on ice to thaw/equilibrate.
- Dissolve 1 protease inhibitor tablet (Roche) in 2 mL water for a 25X Protease Inhibitor stock. After buffer prep, the remaining 25X stock can be stored for 12 weeks at -20˚C.
- Prepare Wash Buffer by combining Pre-Wash Buffer, 25X Protease Inhibitor, and 1 M Spermidine as outlined in Table 1. Store final buffer at RT.
- To a new tube labeled Cell Permeabilization Buffer, add Wash Buffer as outlined in Table 1. Add 5% Digitonin as optimzed for your cell type (see Important Notes on Buffer Prep, above)
- In a new 1.5 mL tube labeled Antibody Buffer, combine Cell Permeabilization Buffer and 0.5 M EDTA as described in Table 1. Place final buffer on ice.
- Store remaining Cell Permeabilization Buffer at 4˚C for use on Day 2.
Table 1. Buffer recipes for CUT&RUN. Includes extra volume to account for pipetting error.
Section II: ConA bead activation (~30 min)
Tips for working with magnetic ConA beads
- Do NOT use ConA beads that have been frozen and/or appear black, granular, or clumpy.
- Do NOT let ConA beads dry out. Avoid disturbing beads with pipette while on magnet.
- Keep activated ConA beads on ice and use within four hours of activation.
- Gently resuspend ConA beads and transfer 11 μL per reaction to a 1.5 mL tube.
- Place tube on a magnet, allow slurry to clear. Pipette to remove supernatant.
- Remove tube from magnet. Immediately add 100 μL per reaction cold Bead Activation Buffer and pipette to resuspend. Return tube to magnet, allow slurry to clear, and pipette to remove supernatant.
- Repeat the previous step one time.
- Resuspend beads in 11 μL per reaction cold Bead Activation Buffer.
- Aliquot 10 μL per reaction of bead slurry into 8-strip tubes. Place on ice.
Section III: Binding cells to activated beads (~30 min)
Guidelines for successful sample prep
- High quality sample prep is essential to CUT&RUN success and is the main variable we see when troubleshooting customer experiments.
- Freshly isolated, native (i.e. unfixed) cells are the preferred input for CUT&RUN. For sample inputs other than native suspension cells (e.g. adherent cells) see Sample Prep.
- Count and examine cells using Trypan Blue staining. Check cellular morphology, integrity, and viability at the three steps outlined below. For more, see this article.
Initial cell harvest. Cells have high viability and expected morphology. Acceptable viability is dependent on cell type and experimental conditions.
Before ConA bead binding. Cells in CUT&RUN Wash Buffer have expected morphology and minimal lysis and/or sample loss.
After binding to ConA beads. The supernatant contains very few cells and the sample shows that all cells are bound to ConA beads.
- Count starting cells and confirm integrity using Trypan Blue staining. Harvest 500,000 cells per reaction (plus 10% excess) and spin at 600 x g for 3 min at room temperature (RT). Remove supernatant.
- Resuspend cells in 100 μL per reaction RT Wash Buffer by gentle yet thorough pipetting. Spin at 600 x g, 3 min, RT. Pipette to remove supernatant.
- Repeat the previous step one time.
- Resuspend cells in 105 μL per reaction RT Wash Buffer. Count and examine integrity of prepared cells using Trypan Blue staining.
- Add 100 µL cells to 10 µL ConA beads in 8-strip tubes. Gently vortex to mix and quick spin in a mini-centrifuge to collect slurry (beads should not settle).
- Incubate bead-cell slurry for 10 min at RT. Cells will adsorb to beads.
- During incubation, retrieve a compatible 8-strip tube magnetic rack. In addition, if using a multi-channel pipettor (recommended), place a multi-channel reagent reservoir on ice. Fill with cold Antibody Buffer.
a. If using the EpiCypher 8-strip tube magnet, use the high-volume side unless otherwise noted. - After the 10 min incubation, place tubes on a magnet and allow slurry to clear. If bead binding was successful, the supernatant should not contain cells. To confirm, save 10 μL supernatant to assess sample integrity using Trypan Blue staining.
- Pipette to remove and discard the remaining supernatant. Remove tubes from magnet and immediately add 50 μL cold Antibody Buffer to each reaction.
- Pipette to resuspend. Confirm ConA bead binding.
Section IV: Antibody binding (~30 min + overnight)
Antibody Incubation Notes
- Add K-MetStat Panel to control reactions BEFORE adding the primary antibody.
- Do NOT rotate or invert tubes. Rotation causes ConA beads to stick to tube sides and dry out, reducing yields. Use a nutator for incubations and elevate tube caps as suggested.
- Quick spin the K-MetStat Panel and pipette to resuspend – do NOT vortex. Add 2 µL K-MetStat Panel to reactions designated for H3K4me3 positive and IgG negative control antibodies. Gently vortex to mix and quick spin tubes. If using fewer than 500,000 cells, decrease K-MetStat Panel amount as in Table 2 (below).
- Add 0.5 μg primary antibody (or manufacturer’s recommendation) to each reaction. For positive and negative control reactions, add 1μL H3K4me3 Positive Control Antibody and 1μL IgG Negative Control Antibody, respectively. For antibodies stored in viscous glycerol solutions, ensure accurate pipetting: aspirate slowly, check tip for accuracy, and pipette up and down to clear the solution from tip.
- Gently vortex reactions to mix. Incubate overnight on a nutator at 4˚C with tube caps elevated (Figure 1). Do NOT rotate – see Antibody Incubation Notes, above.

Figure 1. 8-strip tubes placed on a nutator at a 45 degree angle with caps elevated.
| Cell number | Panel dilution | Volume added to reaction |
| 500,000 | Use stock | 2 μL |
| 250,000 | 1:2 | 2 μL |
| 100,000 | 1:5 | 2 μL |
| 50,000 or fewer | 1:10 | 2 μL |
Table 2. Scale the amount of K-MetStat Panel to the number of cells. For <500,000 cells, prepare a working stock dilution of the K-MetStat Panel in Antibody Buffer the day of experiment.
CUT&RUN Protocol: Day 2
Section V: pAG-MNase binding (~40 min)
Notes on ConA bead-cell clumping
- It is essential that ConA beads remain in solution during pAG-MNase binding and digestion. Excessive bead clumping leads to sample loss, reduces yields, and negatively impacts quality.
Guidelines for high-throughput processing
- Multi-channel pipetting is recommended to improve reliability and experimental throughput. To easily dispense buffers, use multi-channel reagent reservoirs (keep on ice).
- For 8-strip tubes, remove and replace buffers one strip at a time to avoid ConA bead dry out.
- If using a multi-channel pipettor (recommended), place a multi-channel reagent reservoir on ice. Fill with cold Cell Permeabilization Buffer.
- Remove tubes from 4˚C incubation and quick spin to collect liquid. Note that beads may settle overnight (Figure 2, below), but this will not impact results.
- Place tubes on magnet and allow slurry to clear. Pipette to remove supernatant. If using a multi-channel pipettor, remove and replace buffers one tube strip at a time to avoid ConA bead dry-out.
- Keeping tubes on magnet, add 200 μL cold Cell Permeabilization Buffer to each reaction. Pipette to remove supernatant.
- Repeat the previous step one time (keep tubes on magnet).
- Remove tubes from magnet and immediately add 50 μL cold Cell Permeabilization Buffer to each reaction. Gently vortex to mix. Beads may become clumpy at this stage of the protocol, but can be dispersed by gentle pipetting. The end of a pipette tip can be cut off to help mix delicate cells.
- Add 2.5 μL pAG-MNase to each reaction. Gently vortex and/or pipette to thoroughly mix beads and evenly distribute enzyme.
- Incubate reactions for 10 min at RT.
- Quick spin tubes, place on magnet, and allow slurry to clear. Pipette to remove supernatant.
- Keeping tubes on magnet, add 200 μL cold Cell Permeabilization Buffer directly onto beads. Pipette to remove supernatant.
- Repeat one time.
- Remove tubes from magnet. Immediately add 50 μL cold Cell Permeabilization Buffer to each reaction. Gently vortex to mix and disperse clumps by pipetting.

Figure 2. Settling of ConA beads after overnight incubation at 4˚C.
Section VI: Targeted chromatin digestion and release (~3 hrs)
Guidelines for using E. coli spike-in DNA
- Reconstitute the lyophilized E. coli Spike-in DNA prior to first use: Quick spin tube before opening to collect E. coli DNA in bottom of tube. Add 200 μL DNase-free water and thoroughly vortex tube on all sides to resuspend E. coli DNA. Store at -20˚C.
- Aim for E. coli Spike-in DNA to comprise 0.5 – 5% (ideally ~1%) total sequencing reads.
- This protocol is optimized using 0.5 ng E. coli DNA and 500,000 cells per reaction. If using fewer than 500,000 cells per reaction, dilute E. coli Spike-in DNA as outlined here.
- Place tubes on ice. Add 1 μL 100 mM Calcium Chloride to each reaction. Gently vortex and/or pipette to evenly resuspend beads and ensure efficient digestion.
- Incubate tubes on nutator (capped ends elevated) for 2 hours at 4ºC.
- Retrieve E. coli Spike-in DNA. Reconstitute DNA prior to first use (see Guidelines, above) or thaw previously resuspended DNA on ice and quick spin before use.
- Prepare a Stop Master Mix in a 1.5 mL tube. Per reaction, combine 33 μL Stop Buffer and 1 μL E. coli Spike-in DNA (0.5 ng). Gently vortex to mix.
- At the end of the 2 hour incubation, quick spin 8-strip tubes to collect liquid. Add 34 μL Stop Master Mix to each reaction and gently vortex to mix.
- Place reactions in a thermocycler set to 37˚C. Incubate for 10 min.
- Transfer supernatants containing CUT&RUN-enriched DNA to new tubes:
For kit versions 1-3, which include DNA purification spin columns, transfer the supernatants to 1.5 mL tubes. Follow DNA purification protocol in Section VIIb.
For kit versions 4 and later, transfer supernatants to new 8-strip tubes. Follow bead-based purification strategy below (Section VIIa).
Section VIIa: DNA purification (~30 min)
- Prepare 85% Ethanol (EtOH) FRESH using a 100% EtOH stock and molecular biology grade water. Make 500 μL per reaction: 425 μL 100% EtOH + 75 μL water. These calculations include extra volume to account for pipetting error.
- Retrieve SPRIselect reagent, manufactured by Beckman Coulter, Inc., from the kit RT reagents. Vortex thoroughly to resuspend.
- Slowly add 119 μL SPRIselect reagent (1.4X volume) to each reaction. Ensure pipette tip is free of extra droplets before dispensing beads to reactions.
- Gently vortex tubes to mix and quick spin to collect liquid in tube bottom. Incubate 5 min at RT.
- Place tubes on magnet for 2-5 min at RT. Pipette to remove supernatant, being careful not to disturb beads with pipette tip.
- Keeping tubes on magnet, add 180 μL 85% EtOH directly onto beads. Pipette to remove supernatant.
- Repeat the previous step one time.
- Remove tubes from magnet and quick spin, caps facing in. Beads should stay in place on side of tube. Return to magnet and pipette to remove residual EtOH.
- Remove tubes from magnet, leaving caps open. Air-dry beads for 2-3 min at RT, or until liquid has evaporated but beads still appear damp matte brown. If beads become crackly, they are too dry (Figure 3, below).
- Add 17 μL 0.1X TE Buffer to each reaction to elute DNA.
- Gently vortex tubes to resuspend beads and quick spin. Incubate 2 min at RT.
- Place tubes on magnet for 2 min at RT.
- Transfer 15 μL CUT&RUN-enriched DNA to new 8-strip tubes.
- Use 1 μL to quantify DNA using the Qubit fluorometer and 1X dsDNA HS Assay Kit.
Safe pause point.
Review details on confirming CUT&RUN success and continue to library prep or store DNA at -20˚C.
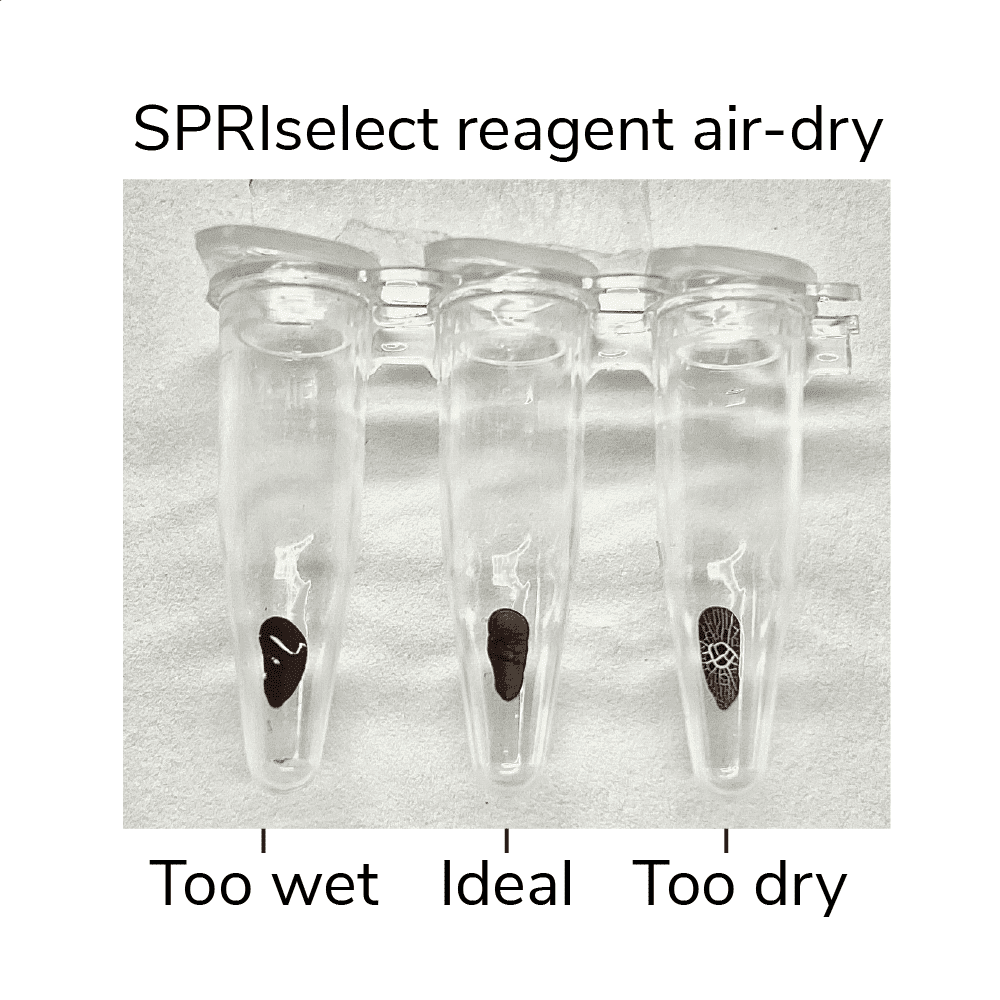
Figure 3. SPRIselect reagent shown at various levels of dry. Ensure your ConA beads are not over or under-dry when eluting DNA.
Section VIIb: DNA purification for Kit Versions 1-3:
Notes before starting DNA purification
- Prior to first use, add 6.9 mL isopropanol to DNA Binding Buffer.
- Prior to first use, add 20 mL ≥95% ethanol to Wash Buffer.
- A vacuum manifold can be used in place of centrifugation. For each step, add the indicated buffer, turn the vacuum on, and allow the solution to pass through the column before turning the vacuum off.
- Add 420 μL DNA Binding Buffer to each reaction. Mix well by vortexing
- For every CUT&RUN reaction, place a DNA Cleanup Column into a DNA Collection Tube. Load each reaction onto a column and label the top.
- Spin at 16,000 x g, 30 sec, RT. Discard flow-through. Place column back into collection tube.
- Add 200 μL DNA Wash Buffer to each column. Spin at 16,000 x g, 30 sec, RT. Discard flow-through. Place column back into collection tube.
- Repeat previous step one time.
- Spin at 16,000 x g, 30 sec, RT to completely dry the column
- Carefully remove the column from the collection tube, ensuring it does not contact the flow-through. Transfer column to a clean pre-labeled 1.5 mL tube.
- Add 12 μL DNA Elution Buffer, taking care to ensure the buffer is added to the center of the column rather than the wall. Tap the column & microfuge collection tube on the benchtop to ensure all droplets are absorbed onto the resin.
DNA can be eluted in 6–20 μL; 12 µL is recommended. Larger elution volumes, longer incubation times, and/or multiple rounds of elution may improve DNA yield. However, DNA concentration will be reduced with larger elution volume. - Let sit 5 minutes, then spin at 16,000 x g, 1 min, RT.
- Vortex eluted material to mix. Use 1 μL to quantify the CUT&RUN DNA using the Qubit fluorometer with the 1X dsDNA HS Assay Kit per the manufacturer’s instructions.
Safe pause point.
Review details on confirming CUT&RUN success and continue to library prep or store DNA at -20˚C.
Confirming CUT&RUN success
- There is no typical yield for CUT&RUN, as results vary by cell type, number, target, and antibody performance.
- In general, yields from H3K4me3 positive control are slightly greater than IgG negative control. However, if yields are similar, this does NOT imply assay failure. Read more here.
- We recommend aiming for ≥5 ng CUT&RUN enriched DNA, which will enable robust library prep. See this article for troubleshooting low yields.
- Electrophoretic analysis (e.g. TapeStation) and/or qPCR of CUT&RUN DNA at this step is NOT recommended.
Here, learn how to go from CUT&RUN-enriched DNA to sequencing. For a streamlined workflow use our CUTANA™ CUT&RUN Library Prep Kit, optimized specifically for CUT&RUN DNA yields.
Looking for the CUTANA CUT&RUN Kit protocol? See this article.
Section VIII: Next-generation sequencing library prep (~4 hrs)
Notes on library prep
- For some cell types/targets, low CUT&RUN DNA yields are unavoidable. To optimize library prep for low DNA inputs, see this article.
- Do NOT shear or fragment DNA before library prep. Our PCR conditions for library prep specifically amplify DNA fragments from 200 to 700 bp, which eliminates large fragments.
- Prepare Illumina® sequencing libraries using ~5 ng purified CUT&RUN DNA and the CUTANA CUT&RUN Library Prep Kit (EpiCypher 14-1001 & 14-1002).
a. For low-abundance targets or if yields <5 ng, use total amount of recovered DNA. Note that IgG and H3K4me3 control antibodies often generate low yields.
b. If using other library prep kits, follow EpiCypher’s recommended PCR parameters for indexing PCR and library amplification (below). These conditions are specifically optimized for small CUT&RUN fragments (200-700 bp).
| Step | Temperature | Time | Cycles | Notes |
| 1. | 98°C | 45 sec | 1 | Hot start activation of DNA Polymerase |
| 2. | 98°C | 15 sec | DNA melting | |
| 3. | 60°C | 10 sec | Hybrid annealing/extension | |
| 4. | Repeat steps 2 and 3 14 times | Amplification | ||
| 5. | 72°C | 60 sec | 1 | Final extension |
| 6. | 4°C | ∞ | Hold |
Section IX: Analysis of library fragment size (~1 hr)
Notes on expected yields and fragment size enrichment
- Fragment distribution analysis of purified sequencing libraries is the single BEST method to confirm CUT&RUN success.
- Libraries should show enrichment of mononucleosome-sized DNA fragments (~300 bp, including CUT&RUN DNA + sequencing adapters). Fragment distributions for positive (e.g. H3K4me3) and negative (e.g. IgG) control reactions can be used to assess yields and validate library prep workflows.
- Final CUT&RUN library concentration is usually 100-200 nM. Libraries ≥1 nM allow pooling at standard concentrations for sequencing, but good data are obtained down to 0.5 nM. If library concentrations are <0.5 nM, read these tips.
- Adapter dimer contamination appears as a peak at ~125 bp and is caused by low input; see this article and the CUTANA CUT&RUN Library Prep Kit Manual for details.
- See this article for troubleshooting low library yields and/or fragment distribution results.
- Use 1 μL purified CUT&RUN library for quantification. Use the Qubit fluorometer with the 1X dsDNA HS Assay Kit per the manufacturer’s instructions.
- For each library, prepare 5 µL at 10 ng/µL for loading onto the Bioanalyzer orTapeStation system. Record the dilution factor, which is needed to calculate library molarity from the results (reported as DNA concentrations in nM for the desired 200 – 700 bp region).
- Load and analyze 1 µL diluted sequencing library using the High Sensitivity DNA Kit (Bioanalyzer) or the D1000 ScreenTape System & Reagents (TapeStation) as per the manufacturer’s instructions.
- The final traces should show predominant enrichment of mononucleosome-sized fragments, such as those yielded by H3K4me3 and CTCF antibodies in Figure 1 (~300 bp: ~170 bp + 125 bp sequencing adapters). Adapter dimers, if present, are observed as a peak at ~125 bp, see this article for more.
Safe pause point. Libraries can be stored at -20˚C for future processing.
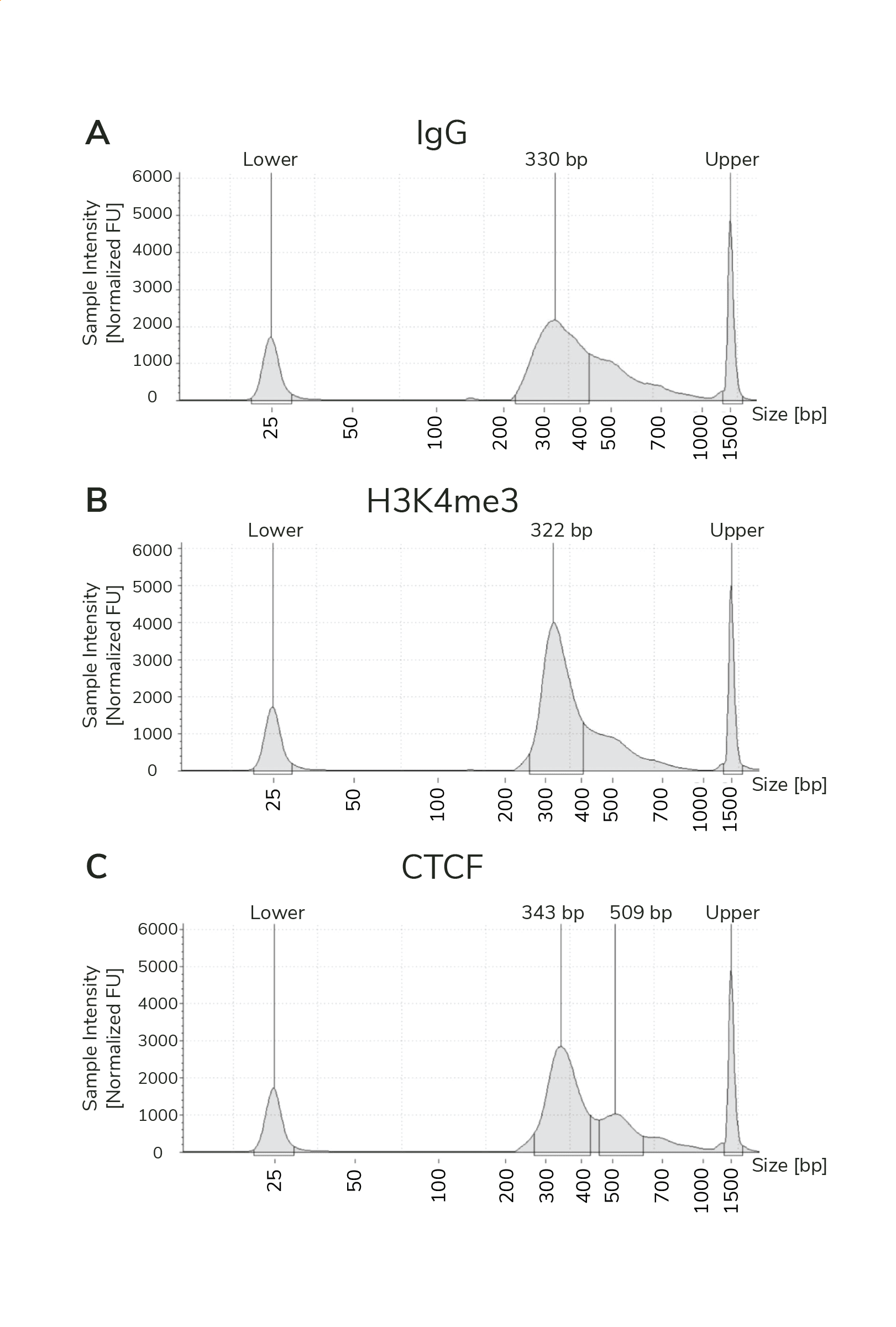
Figure 1. Typical TapeStation traces from CUTANA™ CUT&RUN libraries prepared using antibodies targeting IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0041), and CTCF (EpiCypher 13-2014). All libraries are predominantly enriched for mononucleosome-sized fragments, as indicated by the peak at ~300 bp.
Section X: Illumina sequencing
Tips for sequencing CUT&RUN libraries
- Only 3-8 million uniquely aligned reads are needed for adequate CUT&RUN coverage.
- Paired-end sequencing (2 x 50 bp cycles minimum) is recommended for CUT&RUN to enable detection of K-MetStat Panel barcodes.
- See this article for considerations when sequencing low-concentration libraries.
- See this article for basic information on CUT&RUN sequencing analysis.
- Select appropriate Illumina sequencing platform based on the number of CUT&RUN libraries and desired sequencing depth.
- Pool libraries at desired ratios using the molarity calculations from Section IX (200-700 bp region) and load onto Illumina sequencer. General steps:
a. Confirm that each library in a multiplexed sequencing run has a unique pair of i5 & i7 indexes. Libraries with the same pair of indexes must be sequenced in separate lanes/flow cells.
b. Dilute each library to the same nM concentration, depending on final yields. For NextSeq 2000 and NextSeq 500/550, dilute to 1-4 nM.
c. Pool equimolar libraries into one tube.
d. Dilute pooled libraries to appropriate concentration and in the volume required for Illumina platform. Follow guidelines from specific Illumina kit to load onto sequencer (support.illumina.com).
e. When setting up the sequencing run, make sure dual i5 & i7 indexes are correctly assigned for each library. - For H3K4me3 and IgG control reactions spiked with the K-MetStat Panel, align paired-end sequencing reads to the PTM-specific DNA barcodes. Use this data to validate your workflow, identify failed reactions, and troubleshoot problematic experiments. See this article for guidance.
- If control reactions generate expected results, proceed to analysis of experimental reactions. Align paired-ends reads to the appropriate reference genome (e.g. using Bowtie 2) as described.
- For sequencing normalization using E. coli Spike-in DNA, see this article.
CUTANA CUT&RUN Library Prep Kit
(1) This Product is covered by one or more patents, trademarks and/or copyrights owned or controlled by NEB. While NEB develops and validates its products for various applications, the use of this product may require the purchaser to obtain additional third-party intellectual property rights for certain applications.
(2) This product is licensed for research and commercial use from Bio-Rad Laboratories, Inc., under U.S. Pat. Nos. 6,627,424, 7,541,170, 7,670,808, 7,666,645, and corresponding patents in other countries. No rights are granted for use of the product for Digital PCR or real-time PCR applications, with the exception of quantification in Next Generation Sequencing workflows.
Kit components are stable for 6 months upon date of receipt. Store as outlined below.
Store at room temperature (RT) upon receipt:
| Item | Catalog Number | Notes |
| 8-strip Tubes | 10-0009a | Enables use of multi-channel pipettors. |
| DNA Cleanup Columns *for kit Versions 1-3 | 10-0010 | Use with the DNA Collection Tubes. |
| DNA Collection Tubes *for kit Versions 1-3 | 10-0011 | Use with the DNA Cleanup Columns. |
| DNA Binding Buffer *for kit Versions 1-3 | 21-1008 | Before first use, add 6.9 mL isopropanol. WARNING: Contains toxic ingredients (see Safety Data Sheet). |
| DNA Wash Buffer *for kit Versions 1-3 | 21-1009 | Before first use, add 20 mL of ≥95% ethanol. |
| DNA Elution Buffer *for kit Versions 1-3 | 21-1010 | Use to elute final CUT&RUN DNA. |
| SPRIselect Reagent Manufactured by Beckman Coulter **for kit Version 4 | 21-1405 | DO NOT FREEZE. Reagent is slightly viscous. Thoroughly mix prior to use and pipette carefully to ensure correct volume is transferred. Used to purify CUT&RUN-enriched DNA from assay supernatant. |
| 0.1X TE Buffer **for kit Version 4 | 21-1025 | Used to elute CUT&RUN-enriched DNA. |
| 0.5 M EDTA | 21-1006 | 250X concentration. Use to prepare Antibody Buffer FRESH for each experiment. |
| 100 mM Calcium Chloride | 21-1007 | Activates chromatin-tethered pAG-MNase to cleave DNA. |
Store at 4°C upon receipt:
| Item | Catalog Number | Notes |
| ConA Beads | 21-1401 | DO NOT FREEZE. Concanavalin A (ConA) beads are used for immobilizing nuclei or cells. Because ConA can cause immune cell activation, it is recommended to use nuclei for immune cell studies (see this article). |
| E. coli spike-in DNA | 18-1401 | 100 ng lyophilized E. coli DNA. Before first use, quick spin and reconstitute in 200 μL DNase-free water (0.5 ng/μL). Add to reactions for sequencing normalization. NOTE: After reconstitution, store at -20°C. |
| Bead Activation Buffer | 21-1001 | Use to prepare ConA beads prior to sample immobilization. |
| Pre-Wash Buffer | 21-1002 | Use to prepare Wash, Cell Permeabilization, and Antibody Buffers FRESH for each experiment. |
| Stop Buffer | 21-1003 | 3X concentration. Use to terminate pAG-MNase cleavage activity |
Store at -20°C upon receipt:
| Item | Catalog Number | Notes |
| 5% Digitonin | 21-1004k | Thaw at RT. Use to prepare Cell Permeabilization and Antibody Buffers FRESH for each experiment. Final Digitonin concentration should be optimized for each sample type, see this article. |
| 1 M Spermidine | 21-1005 | 2,000X concentration. Use to prepare Wash Buffer FRESH for each experiment. |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002k | SMALL VOLUME: quick spin before use. Pipette to resuspend – DO NOT VORTEX. Panel of biotinylated nucleosomes coupled to streptavidin-coated magnetic beads. Pair with IgG and H3K4me3 control antibodies. Sufficient for 20 reactions. See this section for more information. |
| Rabbit IgG Negative Control Antibody | 13-0041k | SMALL VOLUME: quick spin before use. 0.5 mg/mL stock. Add 1μL to negative control reactions. Sufficient volume for 10 reactions. |
| H3K4me3 Positive Control Antibody | 13-0042k | SMALL VOLUME: quick spin before use. 0.5 mg/mL rabbit mixed monoclonal antibody. Add 1 μL to positive control reactions. Sufficient volume for 10 reactions. |
| pAG-MNase | 15-1016 | 20X concentration. Proteins A and G (pAG) bind antibodies of various isotypes and host species including total IgG for rabbit, mouse, goat, donkey, rat, guinea pig, horse, and cow. |
Reagents required by not supplied:
- Antibody to target of interest (user-dependent). See this section for more information on antibody selection
- Optional: additional SNAP-CUTANA K-MetStat Panel of spike-in controls, if mapping a target in the K-MetStat Panel
NOTE: The K-MetStat Panel included with the kit is sufficient for positive and negative control reactions only. - Protease inhibitor (e.g. cOmplete™, EDTA-free Protease Inhibitor Cocktail, Roche 11873580001)
- 0.4% Trypan Blue (e.g. Invitrogen T10282)
- Isopropanol (any vendor, for kit Versions 1-3 only)
- 100% Ethanol (any vendor, for kit Version 4)
- Molecular biology grade water, any vendor
- DMSO and PBS, for optimizing Digitonin permeabilization of cells
- CUTANA™ CUT&RUN Library Prep Kit, 48 reactions (EpiCypher 14-1001 & 14-1002)
a. The two versions of this kit contain distinct primer sets, allowing up to 96 CUT&RUN libraries to be multiplexed when kits are used together.
Equipment required but not supplied:
- 1.5, 15 and 50 mL tubes
- Magnetic separation rack for 1.5 mL tubes (e.g. EpiCypher 10-0012) and 8-strip tubes (e.g. EpiCypher 10-0008)
- Qubit™ 4 Fluorometer and 1X dsDNA HS Kit
- 8-channel multi-channel pipettor and multi-channel reagent reservoirs
- Vortex
- Thermocycler
- Tube nutator for incubation stepsa. It is critical to use a nutator rather than a rotator to keep liquid in tube conical bottom and avoid bead drying.
- Capillary electrophoresis machine and required reagents
a. Used AFTER library prep to check the quality of final purified sequencing libraries.
Core CUT&RUN buffer components
| Component |
| HEPES |
| KCl |
| CaCl2 |
| MnCl2 |
| Molecular biology grade water (RNase, DNase free) |
| NaCl |
| EDTA (prepare 0.5 M stock at pH 8.0) |
| EGTA (prepare 0.5 M stock at pH 8.0) |
| RNase A |
| Glycogen |
| Spermidine trihydrochloride* |
| Digitonin (store aliquots of 5% stock in DMSO at -20ºC) |
| DMSO |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail |
| Trypan blue |
*1M spermidine preparation: Dissolve 1 gram spermidine (MW = 254.63) in 3.93 mL molecular grade water. Store in single-use aliquots at -20°C for 6 months.
Core CUT&RUN buffer recipes
| Bead Activation Buffer |
| 20 mM HEPES, pH 7.9 |
| 10 mM KCl |
| 1 mM CaCl2 |
| 1 mM MnCl2 |
| Filter sterilize. Store at 4ºC for up to 6 months. |
| Pre-Wash Buffer |
| 20 mM HEPES, pH 7.5 |
| 150 mM NaCl |
| Filter sterilize. Store at 4ºC for up to 6 months. |
| Wash Buffer |
| Pre-Wash Buffer (recipe above) |
| 0.5 mM Spermidine* |
| 1x Roche cOmplete™, Mini, EDTA-free Protease Inhibitor (CPI-mini, 1 tab/10mL) |
| Filter sterilize. Store at 4ºC for up to 1 week. |
| Antibody Buffer |
| Digitonin Buffer (recipe above) |
| 2 mM EDTA |
| Prepare fresh each day and store at 4ºC. |
| Digitonin Buffer |
| Wash Buffer (recipe above) |
| 0.01% Digitonin** |
| Prepare fresh each day and store at 4ºC. |
| Stop Buffer |
| 340 mM NaCl |
| 20 mM EDTA |
| 4 mM EGTA |
| 50 μg/mL RNase A |
| 50 μg/mL Glycogen |
| Filter sterilize. Store at 4ºC for up to 6 months. |
Buffer preparation notes:
*Spermidine is added to compensate for the removal of Mg2+ from the buffer. Mg2+ can cause DNA degradation and is typically omitted from CUT&RUN buffers.
**Optimal Digitonin concentration for each cell type should be determined empirically, as described. Starting concentration validated for K562, MCF7, and A549 cells is 0.01% digitonin.
Accessory CUT&RUN buffers
| Pre-Nuclei Extraction Buffer |
| 20 mM HEPES, KOH pH 7.9 |
| 10 mM KCl |
| 0.1% Triton X-100 |
| 20% glycerol |
| Filter sterilize. Store at 4˚C for up to 6 months. |
Trypan Blue is a dye commonly used to assess cell viability.
Materials
Trypan Blue solution, 0.4%
Hemocytometer with brightfield/phase microscope or automated cell counter (i.e. Countess™ automated cell counter)
Note on Trypan Blue
- Trypan Blue is toxic to cells. After adding Trypan Blue dye to cells, move quickly to determine cell viability.
Protocol:
- Add 10 µL of 0.4% Trypan Blue to 10 µL washed cells. Pipette to mix.
- Transfer 10 µL of cell-Trypan Blue mix to a counting slide.
- View under brightfield/phase microscope or cell counter.
- Assess cell morphology and viability and troubleshoot as needed.
a. Viable cells are impermeable to Trypan Blue and will exclude the dye (Trypan Blue negative; Figure A).
b. Dead cells are permeable to Trypan Blue and will stain Trypan Blue positive (Figure A).
c. Isolated nuclei will stain Trypan Blue positive (Figure B).
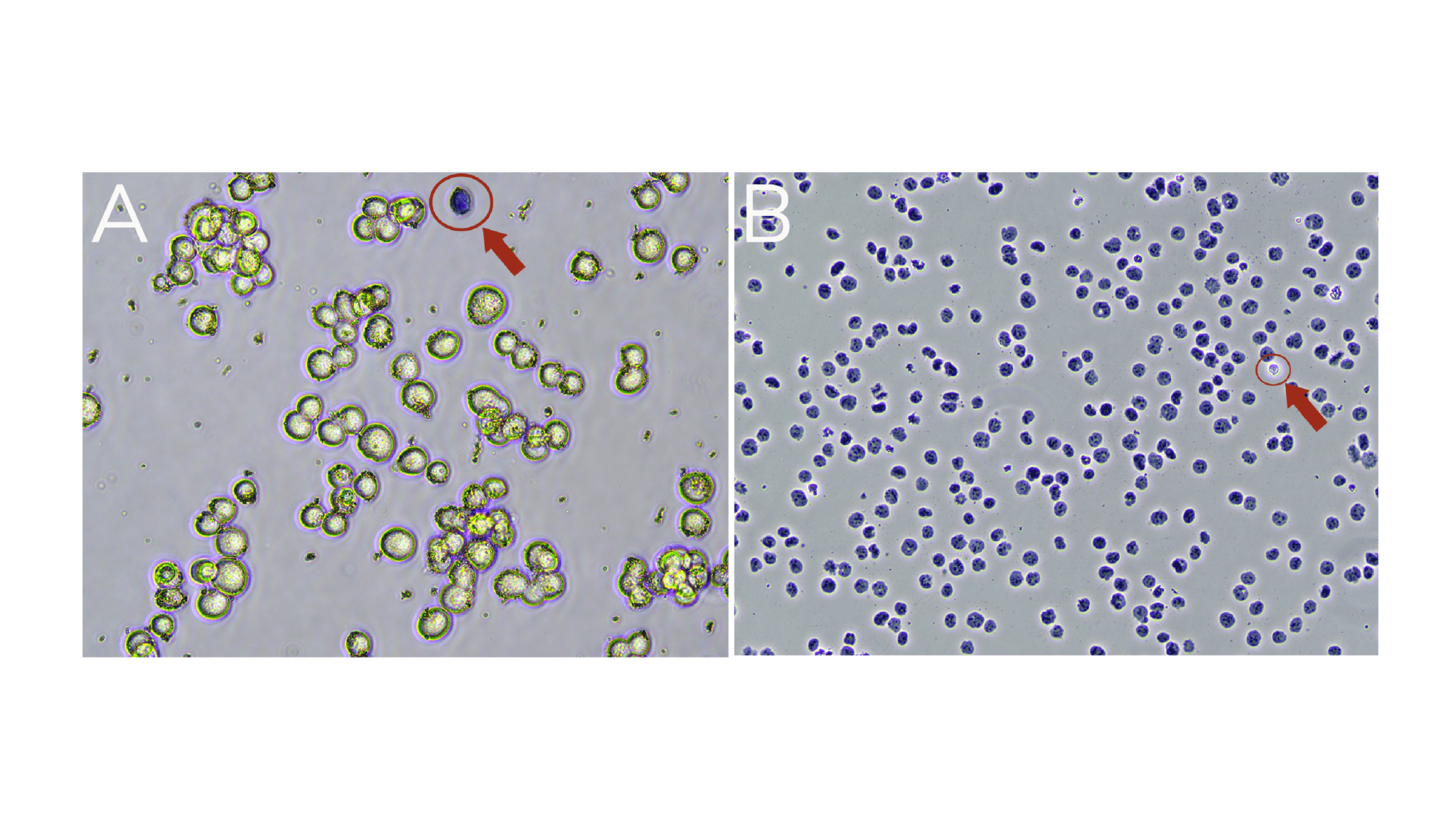
Figure. (A) Washed K562 cells are mostly viable (bright white and round). A dead cell (blue, Trypan positive) is circled in red. (B) Successful nuclei harvest shows Trypan Blue stained nuclei. An intact cell (bright white, Trypan negative) is circled in red.
CUT&RUN uses Digitonin to permeabilize cells and represents a crucial step in the protocol.
Insufficient Digitonin prevents antibody and pAG-MNase from entering the cell, while excess amounts may result in cell lysis. EpiCypher recommends using the minimal amount of Digitonin required to permeabilize >95% of cells. Optimize Digitonin concentrations for each cell type used in CUT&RUN as outlined below.
NOTE: If using nuclei, Digitonin optimization is NOT required. Use 0.01% Digitonin in CUT&RUN buffers to prevent the beads from forming a film on the side of tubes.
Before starting, label your tubes:
- Label five fresh 1.5 mL tubes with percent Digitonin (see Table, below) and a sixth tube as Control.
- Label 6 additional tubes with percent Digitonin or as Control. This second set of tubes will be used for cells.
Prepare buffers
- Prepare a series of five Cell Permeabilization Buffers using 5% Digitonin and CUT&RUN Wash Buffer (see Protocol: Section I), FRESH on the day of use. Add the appropriate volume of Wash Buffer to each tube as outlined in the Table. Add 10 µL 5% Digitonin to the first tube, labeled 0.05%. Vortex to mix.
- Prepare the other four Cell Permeabilization Buffers by serial dilution (see Table). Vortex each buffer to mix and place on ice.
- For the Control buffer, prepare 0.05% DMSO in Wash Buffer.
| Final % Digitonin | 0.05 | 0.01 | 0.001 | 0.0001 | 0.00001 |
| Volume from previous tube (µL) | – | 200 | 100 | 100 | 100 |
| Wash buffer (µL) | 990 | 800 | 900 | 900 | 900 |
| 5% Digitonin (µL) | 10 | – | – | – | – |
Use above Table to prepare serial dilutions of Digitonin.
Permeabilize cells
- Harvest cultured cells for permeabilization testing. To determine the number of cells needed for Digitonin optimization, multiply the number of cells used per CUT&RUN reaction (500,000) x 6.2 (six tubes + 20% excess volume for pipetting errors).
- Spin 600 x g, 3 min, room temperature (RT). Remove supernatant. Resuspend cells in 620 µL RT 1X PBS.
- Aliquot 100 µL cells to the second set of labeled tubes that were set aside for cells.
- Spin cells at 600 x g for 3 min at room temperature (RT). Remove supernatant. Resuspend each cell pellet in 100 µL of the assigned Permeabilization Buffer (or Control) and incubate 10 minutes at RT.
- At the end of the incubation, examine each sample by Trypan Blue staining.
a. In a fresh 1.5 mL tube, mix 10 µL cells + 10 µL 0.4% Trypan blue. Load 10 µL onto a hemacytometer/cell counter slide.
b. Count live (intact, Trypan negative) vs. dead (permeabilized, Trypan positive) cells. Select minimum Digitonin concentration that permeabilizes >95% of cells (example in Figure below).
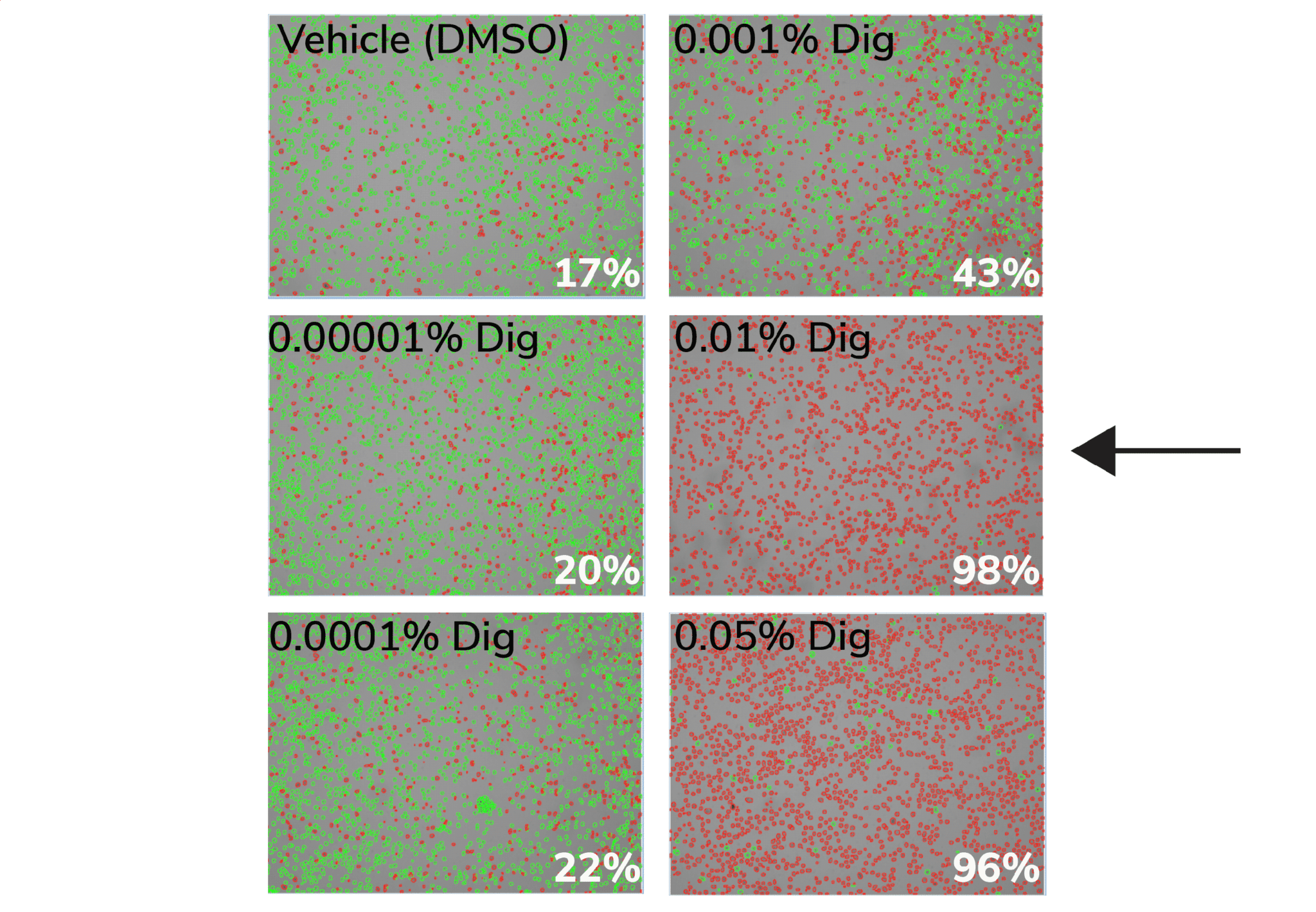
Figure. 0.01% Digitonin is the minimum concentration that permeabilizes >95% K562 cells (black arrow). Cells were treated with CUT&RUN Wash Buffer containing various Digitonin concentrations and evaluated by Trypan Blue staining. Green cells (Trypan negative) are intact, whereas permeabilized/dead cells (Trypan positive) are red. Values (bottom right of each panel) indicate percent of dead/permeabilized cells.
Antibody Selection
CUT&RUN and accessory protocols
Use an antibody validated in CUT&RUN for best chance of success.
For your convenience, EpiCypher offers an array of CUT&RUN certified antibodies for histone post-translational modifications (PTMs) and chromatin associated proteins, all validated in our CUTANA™ CUT&RUN assays. When beginning a CUT&RUN experiment, we recommend first searching our catalog of CUT&RUN validated antibodies.
Don’t see your target? See our articles about CUT&RUN antibody validation for histone PTM or chromatin associated proteins for CUT&RUN, or contact us for recommendations.
You may try, but fair warning: EpiCYpher has found that success in ChIP does NOT guarantee success in CUT&RUN.
This is largely due to differences in sample prep and processing steps. ChIP uses heavily cross-linked cells, stringent wash buffers, and bead-coupled antibodies to help maximize the signal-to-noise ratio. However, these strategies often lead to loss of on-target signal, which is particularly problematic for low abundance targets. To counteract these effect, ChIP requires highly efficient antibodies, with high yields.
In contrast, CUT&RUN uses native chromatin, mild washes, and antibodies in solution, reflecting the increased sensitivity of this newer technique. The only way to know if your antibody will work in CUT&RUN if the antibody has been tested in this assay – antibody validation using ChIP, immunoblot, ELISA, IHC, or other techniques is NOT a predictor of CUT&RUN performance.
For your convenience, EpiCypher offers CUT&RUN-validated antibodies, which you can shop here.
EpiCypher is actively screening antibodies for high-quality performance in CUT&RUN. Visit our antibodies complete list for the most up-to-date list. Below we outline the lot-specific testing criteria for EpiCypher CUT&RUN antibodies across various target classes.
Chromatin-associated protein targets: We offer CUT&RUN antibodies to transcription factors (e.g. CTCF), chromatin reader proteins (e.g. BRD4), modifying enzymes (e.g. MLL1), and remodelers (e.g. SMARCA2, SMARCA4). Each antibody displays high signal-to-noise in CUT&RUN and generates genomic distribution profiles consistent with the reported function of the target protein (for example, DNA binding motif analysis for transcription factors).
Histone PTM targets: Histone PTM antibodies are particularly susceptible to off-target binding, which can compromise biological interpretations. To address these problems, EpiCypher developed the SNAP-CUTANA K-MetStat Panel (EpiCypher 19-1002), and is using these defined nucleosome spike-in controls to identify best-in-class histone lysine methylation PTM antibodies for CUT&RUN. This strategy is the only method that directly confirms antibody specificity in CUT&RUN against physiological on- and off-target substrates. We also validate antibody efficiency, allowing users to be confident when using reduced cell numbers. Each of our SNAP-Certified™ Antibodies show:
- High specificity: <20% recovery of off-target PTMs in the K-MetStat Panel.
- High target efficiency: Robust profiling at 500,000 and 50,000 starting cells.
Using highly specific antibodies will make a huge difference in the biological accuracy and quality of your results. EpiCypher is committed to offering the best histone PTM antibodies for your CUT&RUN studies, so you can be confident in your data. If we do not offer an antibody to your target of interest, contact us for recommendations or take the following steps to validate your own antibody.
- Obtain 3-5 antibodies (preferably monoclonal) to your PTM from various vendors. Make sure that the antibodies target distinct epitopes.
a. EpiCypher scientists have observed that antibodies good for immunofluorescence (IF) applications tend to produce good data in our CUTANA CUT&RUN assays. Although using IF-validated antibodies is NOT a guarantee for success, it may help guide CUT&RUN antibody selection for targets that lack validated reagents - Perform CUT&RUN with all candidate antibodies. Additional controls and recommendations:
a. Include reactions with positive and negative control antibodies (e.g. EpiCypher H3K4me3 SNAP-Certified Antibody and EpiCypher Rabbit IgG Antibody, respectively). Add the SNAP-CUTANA K-MetStat Panel to control reactions to gauge experimental success (see this article).
b. For antibodies to lysine methylation PTMs (H3K4, H3K9, H3K27, H3K36, or H4K20 me1, me2, and me3), add the SNAP-CUTANA K-MetStat Panel to reactions for quantitative antibody validation. See this article for guidance.
c. NOTE: for labile histone PTMs, such as histone lysine acetylation, lightly cross-linking cell samples may stabilize marks and improve assay performance. - Confirm positive and negative controls show expected sequencing results, including data from the SNAP-CUTANA K-MetStat Panel. The negative controls should have low, nonspecific recovery of nucleosomes from the K-MetStat Panel, while the positive control reaction should only recover spike-in nucleosomes carrying the target PTM (e.g. H3K4me3 with less than 20% cross-reactivity to off-targets). Positive controls should also generate robust peaks in expected genomic regions (i.e. sharp peaks at active transcription start sites for H3K4me3). See an expanded discussion and example data here.
- Examine sequencing data and compare the profiles from target antibodies as follows:
a. Lysine methylation PTMs: Antibodies should show <20% cross-reactivity using the SNAP-CUTANA K-MetStat Panel. Confirm that the genomic enrichment is consistent with the target PTM biology (e.g. broad vs narrow peak at functional elements) and shows high signal-to-noise.
b. For other PTMs: Compare results and select a specific antibody based on yields, expected target enrichment, and signal-to-noise in sequencing data.
EpiCypher offers CUT&RUN antibodies to diverse chromatin-associated proteins, including transcription factors (e.g. CTCF), chromatin reader proteins (e.g. BRD4), chromatin modifying enzymes (e.g. MLL1), remodelers (e.g. SMARCA2 and SMARCA4), and commonly used epitope tags (e.g. HA).
Can’t find the antibody you need? Follow the steps below or contact us for recommendations.
We recommend selecting the antibody that best balances the need for robust DNA yields, enrichment for expected sequence motifs and/or peak structures, and high signal over background. Testing native and lightly cross-linked cell samples is also ideal for protein targets.
- Obtain 3-5 antibodies (preferably monoclonal) to your protein from various vendors. Make sure that the antibodies target distinct epitopes.
a. EpiCypher scientists have observed that antibodies good for immunofluorescence (IF) applications tend to produce good data in our CUTANA CUT&RUN assays. Although using IF-validated antibodies is NOT a guarantee for success, it may help guide CUT&RUN antibody selection for targets that lack validated reagents. - Perform CUT&RUN with all candidate antibodies. Additional controls and recommendations:
a. Include reactions with positive and negative control antibodies (e.g. EpiCypher H3K4me3 SNAP-Certified Antibody and EpiCypher Rabbit IgG Antibody, respectively). Add the SNAP-CUTANA K-MetStat Panel to control reactions to gauge experimental success. See this article for guidance. - Confirm positive and negative controls show expected sequencing results, including data from the SNAP-CUTANA K-MetStat Panel. The negative controls should have low, nonspecific recovery of nucleosomes from the K-MetStat Panel, while the positive control reaction should only recover spike-in nucleosomes carrying the target PTM (e.g. H3K4me3 with less than 20% cross-reactivity to off-targets). Positive controls should also generate robust peaks in expected genomic regions (i.e. sharp peaks at active transcription start sites for H3K4me3). See an expanded discussion and example data here.
- Examine sequencing data. Select antibodies that generate high signal-to-noise in CUT&RUN and generate genomic distribution profiles consistent with the reported function of the target protein (for example, DNA binding motif analysis for transcription factors).
Sample Prep
CUT&RUN and accessory protocols
For CUTANA™ CUT&RUN assays, we recommend harvesting 500,000 cells per reaction plus 10-20% excess to account for sample loss.
If your experiment requires fewer cells, see this article for more information.
High-quality sample prep is essential for CUT&RUN experimental success. This guide is for fresh, native (i.e. unfixed, not frozen) suspension cell culture. For alternative sample types (i.e. adherent cultures, tissues, cross-linking, frozen samples), find guidance in this section.
- Count starting cells and confirm cellular integrity, morphology, and viability. It is important that cells have good starting viability, prior to being resuspended in CUT&RUN Wash Buffer. For K652 cells, we aim for >90% viability.
- Harvest 500,000 live cells per reaction plus 10-20% excess. Spin at 600 x g for 3 min at room temperature (RT). Remove supernatant by pipetting, leaving a small amount of liquid on the pellet to avoid sample loss.
- Resuspend cells in 100 µL per reaction RT Wash Buffer by gentle yet thorough pipetting. Spin at 600 x g for 3 min at RT. Pipette to remove supernatant.
- Repeat Step 3 one time.
- Resuspend cells in 105 µL per reaction RT Wash Buffer by gentle yet thorough pipetting.
- Count and examine integrity of prepared cells by Trypan Blue staining.
- Add 100 µL cells to 10 µL ConA beads in 8-strip tubes. Gently vortex to mix and quick spin in a mini-centrifuge to collect slurry (beads should not settle).
- Incubate bead-cell slurry for 10 min at RT to adsorb cells to beads.
- Place tubes on a 8-strip tube magnetic rack and allow slurry to clear.
- If bead binding was successful, the supernatant should not contain cells. Save 10 µL [unbound fraction] to confirm (see Figure 1).
- Discard remaining supernatant and move quickly to the next step. Do not allow beads to dry out.
- Remove tubes from the magnet. Immediately add 50 µL cold Antibody Buffer to each reaction and pipette to resuspend.
- Transfer 10 µL of bead-cell slurry to a new 1.5 mL tube [bead fraction].
- Examine bead-cell slurry [bead fraction] and supernatant [unbound fraction] using Trypan Blue staining to confirm ConA bead binding. As described in Step 10: supernatant should contain few to no cells, bead-cell slurry should contain cells bound to beads (Figure).
Figure 1. (A) Supernatant [unbound fraction] shows little to no material leftover after ConA Bead conjugation. (B) Representative bead-cell slurry [bead fraction] image showing nuclei (blue) successfully conjugated to activated ConA Beads (brown specs). Note: ConA Bead-bound cells will also appear Trypan Blue positive due to the presence of Digitonin in the Antibody Buffer.
High quality sample prep is essential to CUT&RUN success and is the main variable we see when troubleshooting customer experiments. To ensure high quality sample prep, it is essential to examine cellular morphology, integrity, and viability at three steps:
- Initial cell harvest. Cells have high viability and expected morphology.
- Before ConA bead binding. Cells in CUT&RUN Wash Buffer have good integrity, minimal lysis, and/or cell loss.
- After ConA bead binding. Majority of cells are permeabilized and bound to ConA beads.
Initial cell harvest:
This refers to checking cell quality at the beginning CUT&RUN, in Section III of the Protocol. Low starting cell viability, poor morphology, and cellular lysis increase assay background, so it is important to carefully examine each cell type.
Method: Count starting cells and determine viability using our Trypan Blue staining protocol. Examine cell integrity and morphology using a brightfield/phase microscope.
Expected results and troubleshooting:
- Cells should show expected morphology and high viability. For cultured cells, we recommend using cells that are ~70% confluent; do not use overgrown cells.
- Note that viability may vary depending on your cell type (primary vs. cell line), if cells have been treated/stimulated, and other factors. For instance, K562 cells typically show >90% viability, while for other cell types or conditions, the optimal viability may be lower. Make the best decision based your sample type.
Next step in protocol: Harvest 500,000 viable cells per reaction, plus ~10% excess to account for sample loss. If harvesting nuclei, we recommend 10-20% excess.
Before ConA bead binding:
This check is performed just prior to ConA bead binding, when cells are resuspended in Wash Buffer, and represents the final sample quality check before starting CUT&RUN. The purpose of this step is to confirm that washed cells (or nuclei) show normal morphology and remain intact, which is critical for CUT&RUN workflows. Lysed cells are not captured in CUT&RUN.
Method: Determine total cell counts using our Trypan Blue staining protocol. Examine cell integrity and morphology using a brightfield/phase microscope.
Expected results and troubleshooting:
- Cells should show normal morphology/integrity, minimal cell lysis, and be unclumped.
- Cells may show reduced viability in Wash Buffer compared to the initial cell harvest. Instead, focus on total cell counts, confirming ~500,000 cells per reaction.
- Note: if using nuclei, nuclei should all be Trypan Blue positive at this step.
- To troubleshoot cell loss: Increase spin time (keep at 600 x g). Leave ~50 μL liquid on cell pellet when removing supernatant to minimize cell loss, and gently pipette to resuspend.
Next step in protocol: Proceed to ConA bead binding.
After ConA bead binding
This step is to confirm cell permeabilization and ConA bead binding. In Section III of the Protocol, we instruct users to save 10 µL of supernatant following ConA bead binding (unbound fraction) and 10 µL of bead bound sample in Antibody Buffer (bead fraction). Note that Antibody Buffer contains Digitonin, which permeabilizes cells.
Method: Examine unbound fraction and bead fraction using our Trypan Blue staining protocol.
Expected results and troubleshooting:
- Little to no material is present in the unbound fraction (supernatant; Figure A).
- In the bead fraction, >95% of cells (or nuclei) are Trypan Blue positive and surrounded by ConA beads (Figure B).
- To troubleshoot: Ensure ConA beads were never frozen, cells/nuclei were not clumped, beads did not dry out, and all buffers were correctly prepared.
Next step in protocol: Proceed to antibody binding (Section IV).
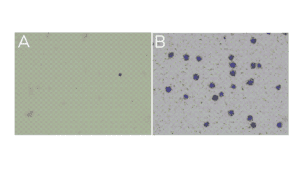
Figure. (A) Unbound fraction has minimal nuclei. (B) Representative sample slurry image showing nuclei (blue) successfully conjugated to activated ConA beads (brown specks).
Collect adherent cells using a mild Trypsin digestion, which dislodges and disaggregates clumps into monodispersed cells without cell damage.
- Incubate with 0.05% Trypsin at 37°C for the minimal time necessary to dislodge cells.
- Add pre-warmed complete media to inactivate Trypsin and then collect cells. Trypsin will be removed during subsequent wash steps.
- Proceed to initial cell count and harvest cells for CUT&RUN.
Concanavalin A (ConA) is a lectin, which can cause immune cell activation. To avoid this potential problem in CUT&RUN, use nuclei or a cross-linking strategy.
Tissues must be processed into a monodispersion of cells, typically by mechanical maceration or douncing. Enzymatic digestion (e.g. collagenase, dispase) can be used for connective tissue and Trypsin may be used for macro-dissected tissues (monitor dissolution to single cells). Harvesting viable, monodispersed cells from tissues can be challenging; in some cases nuclei may be preferable. See literature, including the following papers, for additional methods:
- Carpenter et al. Cell-type specific profiling of histone post-translational modifications in the adult mouse striatum. Nature Communications 13 (2022).
- de Bock et al. HOXA9 cooperates with activated JAK/STAT signaling to drive leukemia development. Cancer Discov 8, 616-631 (2018).
- Janssens et al. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin 11 (2018).
- Larsen et al. Establishment, maintenance, and recall of inflammatory memory. Cell Stem Cell 28, 1758-1774 (2021).
- Liu et al. Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell 173, 430-442 (2018).
- Miao et al. Glucose dissociates DDX21 dimers to regulate mRNA splicing and tissue differentiation. Cell 186, 80-97 (2023).
- Uyehara & McKay. Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. Proc Natl Acad Sci USA 116, 9893-9902 (2019).
Use conditions that minimize lysis, which can contribute to elevated background. Ensure Digitonin is optimized for cell types.
Freezing cells
- Count cells and confirm viability, integrity, and morphology using Trypan Blue staining. Spin cells 600 x g, 3 min, room temperature (RT).
- Remove supernatant. Resuspend in cell culture media with 10% DMSO and aliquot as desired. EpiCypher typically aliquots 5 million cells for 8 reactions, which allows for ~20% sample loss during freeze/thaw.
- Slowly freeze aliquots (-1˚C per minute) in an isopropanol-filled chiller in a -80°C freezer (e.g. “Mr. Frosty”).
Thawing cells
- When ready to perform CUT&RUN, remove tubes from -80˚C and quickly place on a 37°C block to thaw. Work quickly to avoid cell lysis.
- When cells are almost thawed, remove from 37˚C and pipette to fully thaw cells.
- Spin cells at 600 x g, 3 min, RT. Pipette to remove supernatant.
- Resuspend cells in 105 μL per reaction RT Wash Buffer. Take a 10 μL aliquot to count using Trypan Blue staining. Note that viability may be decreased; focus instead on cell integrity, lysis levels, and total cell counts. If significant sample loss has occurred, spin cells again and resuspend in a smaller volume of Wash Buffer.
- Continue to ConA bead binding (Protocol: Section III).
| Materials needed | Source |
| Pre-Nuclei Extraction Buffer | EpiCypher 21-1026a |
| 1M Spermidine | EpiCypher 21-1026b |
| Protease Inhibitor | Roche 11873580001 |
| Phosphate Buffered Saline (PBS) | Any vendor |
| 0.4% Trypan Blue | Invitrogen T10282 |
| Brightfield or phase microscope + hemacytometer slides | Any vendor |
Nuclei Prep Protocol
- Prepare the Nuclei Extraction Buffer fresh on the day of nuclei harvest. In a clean tube, add 250 µL Pre-Nuclei Extraction Buffer per reaction. Add 1 M Spermidine at a 1:2,000 dilution. Then add Protease Inhibitor at a 1X final concentration. Place final Nuclei Extraction Buffer on ice. Note: users may choose to prepare their own Pre-Nuclei Extraction Buffer following this recipe.
- Counts cells and confirm starting cell integrity, morphology, and viability by Trypan Blue staining. It is important that cells have good starting viability. For K652 cells, we aim for >90% viability.
- Harvest 500,000 cells per reaction plus 10-20% excess to account for sample loss.
- Spin at 600 x g for 3 min at room temperature (RT). Remove supernatant and resuspend cells in 100 µL per reaction cold Nuclei Extraction Buffer.
- Incubate on ice for 10 min.
- Spin at 600 x g for 3 min at 4ºC. Remove and discard supernatant. The pellet should change in appearance from sticky, pale yellow (cells) to white and fluffy (nuclei).
- Gently resuspend nuclei in 105 µL per reaction cold Nuclei Extraction Buffer (i.e. for 8 reactions, resuspend in 840 µL).
- Take a 10 µL aliquot to examine nuclear integrity by Trypan Blue staining.
View under brightfield/phase microscope or cell counter to confirm integrity. Isolated nuclei will stain blue, while cells will be bright white and round (see Figure below). - If conducting experiments immediately, continue to Protocol: Section III. Otherwise, see crypreservation protocol below.
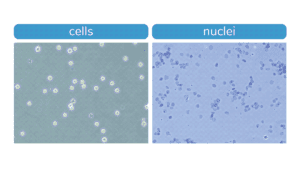
Figure. Morphology characteristic of intact K562 cells (left) compared to isolated nuclei (right) when visualized under brightfield microscope after Trypan Blue staining. Isolated nuclei will stain blue, while cells will be bright white and round. For accurate nuclei counts, record “dead” cell numbers on an automated cell counter or manually count blue stained nuclei.
Nuclei Cryopreservation and Thawing Protocol
- Aliquot nuclei resuspended in Nuclei Extraction Buffer. EpiCypher typically aliquots for >8 reactions, plus 20-30% excess to account for sample loss.
- Slowly freeze aliquots in an isopropanol-filled chiller in a -80°C freezer.
- When ready to use samples for CUT&RUN, thaw nuclei quickly by placing on 37ºC block. Move quickly to avoid nuclear lysis and chromatin fragmentation.
- Thawed nuclei in Nuclei Extraction Buffer can be directly added to activated ConA beads (Protocol: Section III).
CUT&RUN uses Digitonin to permeabilize cells and represents a crucial step in the protocol.
Insufficient Digitonin prevents antibody and pAG-MNase from entering the cell, while excess amounts may result in cell lysis. EpiCypher recommends using the minimal amount of Digitonin required to permeabilize >95% of cells. Optimize Digitonin concentrations for each cell type used in CUT&RUN as outlined below.
NOTE: If using nuclei, Digitonin optimization is NOT required. Use 0.01% Digitonin in CUT&RUN buffers to prevent the beads from forming a film on the side of tubes.
Before starting, label your tubes:
- Label five fresh 1.5 mL tubes with percent Digitonin (see Table, below) and a sixth tube as Control.
- Label 6 additional tubes with percent Digitonin or as Control. This second set of tubes will be used for cells.
Prepare buffers
- Prepare a series of five Cell Permeabilization Buffers using 5% Digitonin and CUT&RUN Wash Buffer (see Protocol: Section I), FRESH on the day of use. Add the appropriate volume of Wash Buffer to each tube as outlined in the Table. Add 10 µL 5% Digitonin to the first tube, labeled 0.05%. Vortex to mix.
- Prepare the other four Cell Permeabilization Buffers by serial dilution (see Table). Vortex each buffer to mix and place on ice.
- For the Control buffer, prepare 0.05% DMSO in Wash Buffer.
| Final % Digitonin | 0.05 | 0.01 | 0.001 | 0.0001 | 0.00001 |
| Volume from previous tube (µL) | – | 200 | 100 | 100 | 100 |
| Wash buffer (µL) | 990 | 800 | 900 | 900 | 900 |
| 5% Digitonin (µL) | 10 | – | – | – | – |
Use above Table to prepare serial dilutions of Digitonin.
Permeabilize cells
- Harvest cultured cells for permeabilization testing. To determine the number of cells needed for Digitonin optimization, multiply the number of cells used per CUT&RUN reaction (500,000) x 6.2 (six tubes + 20% excess volume for pipetting errors).
- Spin 600 x g, 3 min, room temperature (RT). Remove supernatant. Resuspend cells in 620 µL RT 1X PBS.
- Aliquot 100 µL cells to the second set of labeled tubes that were set aside for cells.
- Spin cells at 600 x g for 3 min at room temperature (RT). Remove supernatant. Resuspend each cell pellet in 100 µL of the assigned Permeabilization Buffer (or Control) and incubate 10 minutes at RT.
- At the end of the incubation, examine each sample by Trypan Blue staining.
a. In a fresh 1.5 mL tube, mix 10 µL cells + 10 µL 0.4% Trypan blue. Load 10 µL onto a hemacytometer/cell counter slide.
b. Count live (intact, Trypan negative) vs. dead (permeabilized, Trypan positive) cells. Select minimum Digitonin concentration that permeabilizes >95% of cells (example in Figure below).

Figure. 0.01% Digitonin is the minimum concentration that permeabilizes >95% K562 cells (black arrow). Cells were treated with CUT&RUN Wash Buffer containing various Digitonin concentrations and evaluated by Trypan Blue staining. Green cells (Trypan negative) are intact, whereas permeabilized/dead cells (Trypan positive) are red. Values (bottom right of each panel) indicate percent of dead/permeabilized cells.
While cross-linking is not necessary for CUT&RUN, it may be beneficial for:
- Labile targets (such as histone lysine acetylation).
- Experiments with tightly controlled time points.
- Transiently chromatin-interacting proteins (such as acetyl-lysine reader proteins or remodeling enzymes).
In this article, you will find guidance for incorporating cross-linking into your CUT&RUN experiment. Importantly, ALWAYS include native samples when testing cross-linking conditions.
When optimizing cross-linking conditions:
- Start with light cross-linking (0.1% formaldehyde, 1 min), which generally preserves signal without negatively impacting data.
- If light cross-linking is not sufficient, moderate cross-linking (1% formaldehyde, 1 min) can be attempted with the caveat that it may reduce DNA yield.
- Avoid heavy cross-linking conditions used for ChIP (>1% formaldehyde, 1-10 min) which is deleterious to both DNA yield and data quality.
| Materials needed |
| Pre-Wash Buffer |
| Triton X-100, 10% solution |
| SDS, 10% solution |
| 37% Formaldehyde |
| Glycine |
| 20 μg/μL Proteinase K |
CROSS-LINKING PROTOCOL
CUT&RUN DAY 1
- Prepare CUT&RUN Wash, Cell Permeabilization, and Antibody Buffers using Pre-Wash Buffer supplemented with 1% Triton X-100 and 0.05% SDS.
- Perform cross-linking at the beginning of Protocol: Section III as follows:
a. For suspension culture cells, make sure cells are well mixed and take a 10 µL aliquot to count using Trypan Blue staining. Transfer 500,000 cells per reaction (plus 10% excess) into a 15 mL or 1.5 mL tube.
b. For adherent cells, cross-linking will be performed while cells are still attached to the plate. - Add fresh 37% Formaldehyde directly to culture for a final concentration of 0.1-1%. Test a range of concentrations to optimize for target and cell type.
- Quickly vortex (suspension cells) or swirl plate (adherent cells) to mix.
- Incubate for 1-10 min at room temperature (RT). 1 min is recommended. Test a range of times to determine optimal fixation conditions.
- Quench cross-linking by adding Glycine to a final concentration of 125 mM. Vortex (suspension cells) or swirl (adherent cells) to mix.
a. Suspension cells: Proceed to Protocol: Section III Step 2 of the CUT&RUN protocol (spin at 600 x g for 3 min at RT).
b. For adherent cells: See this article for instructions.
CUT&RUN DAY 2
- Following the collection of supernatants containing CUT&RUN-enriched DNA (Protocol: Section VI), it is crucial to reverse cross-links.
- Add 0.8 μL 10% SDS followed by 1 μL of 20 μg/μL Proteinase K to each supernatant. Vortex to mix and quick spin to collect liquid.
- Place supernatants (in 8-strip tubes) in a thermocycler set to 55˚C. Incubate overnight.
- The next day, quick spin tubes and resume CUT&RUN at Section VII: DNA Purification (columns or beads).
Quality Control Checks
Success metrics and expected results
The CUTANA™ CUT&RUN Kit contains multiple quality control metrics and checks to ensure successful chromatin profiling. Quality control metrics are listed for each section of the CUT&RUN workflow (see Figure, below). For more, navigate to:
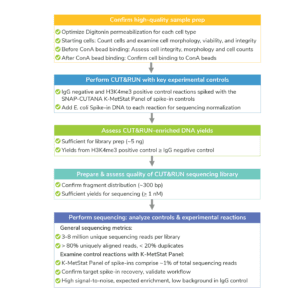
Figure. Overview of CUTANA CUT&RUN success metrics.
High quality sample prep is essential to CUT&RUN success and is the main variable we see when troubleshooting customer experiments. To ensure high quality sample prep, it is essential to examine cellular morphology, integrity, and viability at three steps:
- Initial cell harvest. Cells have high viability and expected morphology.
- Before ConA bead binding. Cells in CUT&RUN Wash Buffer have good integrity, minimal lysis, and/or cell loss.
- After ConA bead binding. Majority of cells are permeabilized and bound to ConA beads.
Initial cell harvest:
This refers to checking cell quality at the beginning CUT&RUN, in Section III of the Protocol. Low starting cell viability, poor morphology, and cellular lysis increase assay background, so it is important to carefully examine each cell type.
Method: Count starting cells and determine viability using our Trypan Blue staining protocol. Examine cell integrity and morphology using a brightfield/phase microscope.
Expected results and troubleshooting:
- Cells should show expected morphology and high viability. For cultured cells, we recommend using cells that are ~70% confluent; do not use overgrown cells.
- Note that viability may vary depending on your cell type (primary vs. cell line), if cells have been treated/stimulated, and other factors. For instance, K562 cells typically show >90% viability, while for other cell types or conditions, the optimal viability may be lower. Make the best decision based your sample type.
Next step in protocol: Harvest 500,000 viable cells per reaction, plus ~10% excess to account for sample loss. If harvesting nuclei, we recommend 10-20% excess.
Before ConA bead binding:
This check is performed just prior to ConA bead binding, when cells are resuspended in Wash Buffer, and represents the final sample quality check before starting CUT&RUN. The purpose of this step is to confirm that washed cells (or nuclei) show normal morphology and remain intact, which is critical for CUT&RUN workflows. Lysed cells are not captured in CUT&RUN.
Method: Determine total cell counts using our Trypan Blue staining protocol. Examine cell integrity and morphology using a brightfield/phase microscope.
Expected results and troubleshooting:
- Cells should show normal morphology/integrity, minimal cell lysis, and be unclumped.
- Cells may show reduced viability in Wash Buffer compared to the initial cell harvest. Instead, focus on total cell counts, confirming ~500,000 cells per reaction.
- Note: if using nuclei, nuclei should all be Trypan Blue positive at this step.
- To troubleshoot cell loss: Increase spin time (keep at 600 x g). Leave ~50 μL liquid on cell pellet when removing supernatant to minimize cell loss, and gently pipette to resuspend.
Next step in protocol: Proceed to ConA bead binding.
After ConA bead binding
This step is to confirm cell permeabilization and ConA bead binding. In Section III of the Protocol, we instruct users to save 10 µL of supernatant following ConA bead binding (unbound fraction) and 10 µL of bead bound sample in Antibody Buffer (bead fraction). Note that Antibody Buffer contains Digitonin, which permeabilizes cells.
Method: Examine unbound fraction and bead fraction using our Trypan Blue staining protocol.
Expected results and troubleshooting:
- Little to no material is present in the unbound fraction (supernatant; Figure A).
- In the bead fraction, >95% of cells (or nuclei) are Trypan Blue positive and surrounded by ConA beads (Figure B).
- To troubleshoot: Ensure ConA beads were never frozen, cells/nuclei were not clumped, beads did not dry out, and all buffers were correctly prepared.
Next step in protocol: Proceed to antibody binding (Section IV).

Figure. (A) Unbound fraction has minimal nuclei. (B) Representative sample slurry image showing nuclei (blue) successfully conjugated to activated ConA beads (brown specks).
Analyze CUT&RUN-enriched DNA prior to library prep. Consider the following:
DNA yield
There is no typical DNA yield for CUT&RUN, as yields can vary by cell type, number of cells, target abundance, and antibody quality. Instead, we suggest to:
- Check that yields from the H3K4me3 control are similar to or slightly greater than the IgG negative control.
a. Note that similar and/or low yields from the positive control reflect the low abundance of H3K4me3 and do NOT imply assay failure. Read more here.
b. If a positive control with higher yields is needed, we recommend H3K27me3 (EpiCypher 13-0055). - Aim for ≥5ng CUT&RUN-enriched DNA, which will enable robust library prep.
a. Yields below 5 ng are common for low abundance targets, such as H3K4me3. In these cases, we suggest using all of the CUT&RUN DNA for library prep.
b. For help with low yields, see this FAQ. Note that sometimes low yields cannot be avoided, but modifications can be made to library prep to increase yields for sequencing.
Fragment analysis
Do NOT assess fragment size distribution of CUT&RUN DNA. Raw CUT&RUN yields are too low for detection on Bioanalyzer/TapeStation, and will not provide useful information at this step. Wait until after library prep. For further reading, see this FAQ.
Fragment distribution of purified library
- Fragment distribution analysis of purified sequencing libraries is the single BEST method to confirm CUT&RUN success.
- Libraries should show enrichment of mononucleosome-sized DNA fragments (~300 bp, including CUT&RUN DNA + sequencing adapters) (see Figure 1 below).
Library yields
- In general, library yields should not be used to determine assay success. Library yields vary widely by cell type, number of cells, target abundance, and antibody quality.
- Aim for a library concentration of ≥ 1 nM, which will enable pooling at standard concentrations for multiplexed sequencing.
- For library concentrations below 0.5 nM, we recommend visiting considerations for library prep and sequencing from low CUT&RUN yields and see additional tips here for experiments that cannot be repeated.

Figure 1. Typical TapeStation traces from CUTANA™ CUT&RUN libraries prepared using antibodies targeting IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0041), and CTCF (EpiCypher 13-2014). All libraries are predominantly enriched for mononucleosome-sized fragments, as indicated by the peak at ~300 bp (~170 bp nucleosomes + sequencing adapters).
CUT&RUN sequencing data metrics
- Libraries should be sequenced to a depth of 3-8 million reads. Majority of reads (>80%) should align uniquely to the species genome.
- Sequence duplication levels should be low (<20% of total sequence reads).
- The SNAP-CUTANA™ K-MetStat Panel should comprise ~1% of unique reads and produce expected results in H3K4me3 and IgG control reactions. See this article for further reading on using SNAP-CUTANA Spike-in Controls.
- H3K4me3 and IgG controls should show expected enrichment and peak structures. Experimental replicates should be highly reproducible (Figure).
- E. coli Spike-in DNA should comprise ~1% of total unique reads
- Target enrichment and peak structure should be consistent with biological function (if known).
Looking for more on sequencing analysis?
For help with CUT&RUN sequencing analysis, including genomic alignment, peak calling, and signal-to-noise calculations, see this section.
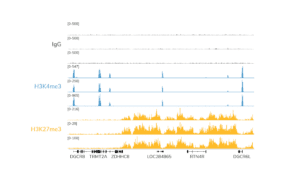
Figure. Data generated by three independent users demonstrate expected enrichment, peak structure, and reproducibility of data generated with the CUTANA™ CUT&RUN kit. CUT&RUN was performed using 500,000 K562 cells and antibodies to IgG (negative control), H3K4me3 (positive control), and H3K27me3. 3-6 million reads were generated per library. H3K4me3 tracks show sharp peaks localized to transcription start sites (TSSs), while H3K27me3 tracks show broad peaks over repressed regions. IgG shows typical low background.
In the CUTANA™ CUT&RUN Kit, EpiCypher adds the SNAP-CUTANA™ K-MetStat Panel to positive (H3K4me3) and negative (IgG) control reactions to confirm workflows and guide troubleshooting. Each CUT&RUN kit comes with sufficient controls antibodies and the K-MetStat Panel sufficient for 10 experiments:
- 10 reaction volumes of IgG Negative Control Antibody
- 10 reaction volumes of H3K4me3 Positive Control Antibody
- 20 reaction volumes of the SNAP-CUTANA K-MetStat Panel (added to both positive and negative control reactions)
Of note, EpiCypher often uses H3K27me3 as an additional positive control (EpiCypher 13-0055), as it has higher yields than H3K4me3. The K-MetStat Panel should also be added to H3K27me3 control reactions; purchase additional K-MetStat Panel here.
Why are these controls useful?
SNAP-CUTANA Spike-in controls are the only control that replicates the in vivo target of CUT&RUN (i.e. nucleosomes). The spike-ins are added to cells just after ConA bead binding and are processed alongside the sample throughout the CUT&RUN protocol (Figure 1). By combining these spike-ins with our rigorously validated CUT&RUN antibodies for H3K4me3 and IgG, we can immediately gauge workflow success.
EpiCypher includes these control reactions in every experiment, and it has saved us countless time and resources in protocol optimization. Should you reach out to our technical support team for troubleshooting assistance, they will be able to assist you much better if you include data derived from this panel. For details on how to leverage the K-MetStat Panel for troubleshooting, see this article.
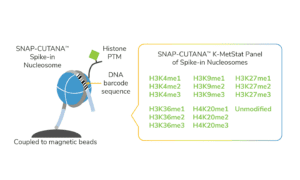
Figure 1. The K-MetStat Panel is composed of 16 spike-in nucleosomes, representing 15 distinct histone lysine methylation PTMs and an unmodified control. Nucleosomes are coupled to magnetic beads for easy one-step addition to CUT&RUN. A PTM-specific DNA barcode enables detection of spike-ins in sequencing data.
Pairing the K-MetStat Panel with control antibodies to determine assay success
We review the specifics of how SNAP-CUTANA Spike-ins are processed in CUT&RUN reactions here; the strategy using the K-MetStat Panel with control antibodies is outlined in Figure 2. Briefly:
- Add the SNAP-CUTANA K-MetStat Panel to designated control reactions immediately prior to the addition of H3K4me3 or IgG control antibody (Figure 2).
- Add antibody, which binds its target in cells and in the spike-in panel (Figure 2).
- pAG-MNase cleaves antibody-bound chromatin and antibody-bound spike-in. Cleaved DNA is purified and prepared for sequencing.
- Samples are sequenced. For each control reaction, download sequencing files and determine the number of sequencing reads aligned to each PTM-specific DNA barcode, using the analysis procedure as described in this article. Barcode read counts provide a useful measurement of PTM recovery and workflow success.
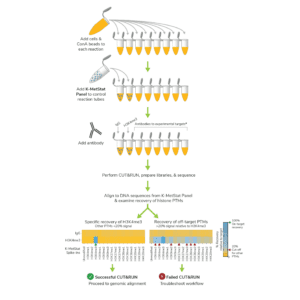
Figure 2. The K-MetStat Panel is used with control antibodies to determine CUT&RUN success. Two different outcomes are shown. Note that the K-MetStat Panel can be added to experimental reactions targeting a PTM in the Panel. Additional K-MetStat Panel can be purchased at 19-1002.
What are the expected results? How is assay success determined?
Figure 2 illustrates two potential outcomes from H3K4me3 and IgG control reactions: one successful CUT&RUN experiment and one that indicates a failed experiment. In each heatmap, reactions are separated into rows and recovery of each PTM in the Panel is separated by column. The heatmap shows data normalized to the on-target PTM (i.e. the on-target PTM is set to 100%, appearing blue). Off-target PTMs should have low signal relative to this target and appear orange. EpiCypher always aims for off-target PTMs to show <20% signal relative to the target PTM.
To determine successful CUT&RUN, keep the following metrics in mind:
- The IgG negative control should show no preference among PTMs and low background.
- H3K4me3 positive control should have strong enrichment for H3K4me3 spike-ins, less than 20% off-target PTM recovery, and high signal-to-noise.
- Spike-in barcode reads should comprise ~1% (0.5-5%) of total sequencing reads. This may vary based on target abundance and sequencing depth. The main goal is to have thousands of reads, which will allow adequate sampling of the K-MetStat Panel for reliable data analysis.
- If control reactions generate expected spike-in data, you can be confident in the technical aspects of your workflow (Figure 2, left heatmap).
- If more than 20% off-target PTM recovery in H3K4me3 control and/or high background in IgG control indicate experimental problems (Figure 2, right heatmap). See this article for a discussion of troubleshooting using spike-in results.
Data Analysis
How to analyze CUT&RUN data
CUT&RUN analysis methods are similar to those used for ChIP-seq datasets, with a few key differences. Briefly:
- Align raw reads to a reference genome using Bowtie2 [1]. The Integrative Genomics Viewer (IGV) [2] and/or deepTools2 [3] can be used to visualize enrichment (e.g. bigWig files graphed over a genome browser).
- For peak calling, EpiCypher frequently uses MACS2 [4] and SICER [5], programs for ChIP-seq that work well for CUT&RUN [6]. SICER can be adjusted for analysis of sharp enrichment peaks (e.g. H3K4me3) vs. broad areas of enrichment (e.g. H3K27me3) [7]. Other options include SEACR [8], a peak caller designed for CUT&RUN data, and the CUT&RUNTools 2.0 pipeline, which is designed for CUT&RUN and CUT&Tag data, including analysis of single cells [9]. It is recommended to test several programs and select the one that faithfully represents the target of interest.
- To determine signal over background, EpiCypher uses bedTools to calculate fractions of reads in peaks (FRiP) and compare FRiP scores from experimental samples vs. controls [10]. Other tools can be applied for differential analysis and heatmap generation (e.g. DESeq2 [11], deepTools2 [3]).
References:
- Langmead & Salzberg. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357-359 (2012).
- Robinson et al. Integrative Genomics Viewer. Nat Biotechnol 29, 24–26 (2011).
- Ramírez et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 8, 44 (2016).
- Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol 1150, 81-95 (2014).
- Zang C et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952-1958 (2009).
- Evans et al. Ybx1 fine-tunes PRC2 activities to control embryonic brain development. Nat Commun 11, 4060 (2020).
- Laczik M et al. Iterative Fragmentation Improves the Detection of ChIP-seq Peaks for Inactive Histone Marks. Bioinform Biol Insights 10, 209-224 (2016).
- Meers et al. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42 (2019).
- Yu F et al. CUT&RUNTools 2.0: A pipeline for single-cell and bulk-level CUT&RUN and CUT&Tag data analysis. Bioinformatics 38, 252-254 (2021).
- Schep AN et al. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14, 975-978 (2017).
- Love MI et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014).
Aim for E. coli Spike-in DNA to comprise ~1% (0.5-5%) of total sequencing reads. In the protocol, 0.5 ng is recommended for 500,000 cells. Generally, this can be decreased linearly with decreasing cell number (e.g. 0.1 ng per 100,000 cells). The amount may need to be adjusted to achieve read counts in the optimal range due to variables such as target abundance, antibody efficiency, etc.
To normalize sequencing results using E. coli Spike-in DNA:
- Align sequencing reads to reference genome (e.g. human, mouse), and filter out muti-mapping reads, reads assigned to blacklisted regions, and duplicate reads (as desired) to determine the total number of uniquely aligned reads. Perform for each reaction.
- In a separate alignment, align sequencing reads to the E. coli K12, MG1655 reference genome: https://support.illumina.com/sequencing/sequencing_software/igenome.html. Filter out reads that do NOT align uniquely. Note that this alignment is separate from the experimental reference genome in Step 1.
- For pairwise comparisons, quantify E. coli Spike-in DNA reads for each CUT&RUN reaction and normalize to the total number of uniquely aligned reads.
Example: CUT&RUN was used to map H3K4me3 in treated and untreated cells.
a. Treatment spike-in = 100,000 E. coli reads in 5,000,000 total reads = 2%
b. Untreated spike-in = 30,000 E. coli reads in 3,000,000 total reads = 1% - Calculate normalization factor (see [1]) such that after normalization the E. coli spike-in signal is set to be equal across all reactions.
Example from above, comparing H3K4me3 in treated vs. untreated cells:
a. Treatment normalization factor = 1 / 2% spike-in bandwidth = 0.5
b. Untreated normalization factor = 1 / 1% spike-in bandwidth = 1.0 - Use single scalar normalization ratio with the — scaleFactor option enabled in deepTools bamCoverage tool to generate normalized bigwig files for visualization in IGV (https://deeptools.readthedocs.io/en/develop/content/tools/bamCoverage.html).
Continuing with the Example from above:
a. Treatment sample –scaleFactor = 0.5
b. Untreated sample –scaleFactor = 1.0
The effect of normalization on a dataset is inversely proportional to the E. coli Spike-in bandwidth. In other words, reactions with the highest bandwidth will receive the largest reduction in signal after normalization. For further information on sequencing normalization using exogenous spike-in controls, see [1,2].
References:
- Tay et al. Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J Exp Med 217 (2020).
- Orlando et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9, 1163-1170 (2014).
How to analyze SNAP-CUTANA™ Spike-in Controls
- Download R1 & R2 paired-end sequencing files (fastq.gz) for control reactions. Double-click the fastq.gz files to create fastq files and save in a new folder.
- On the SNAP-CUTANA Spike-in product page (e.g. K-MetStat Panel), under Documents and Resources, download the Shell Script (.sh) and K-MetStat Panel Analysis (.xlsx) files. Save to the folder from Step 1.
- Open the .sh file in TextEdit or any text editing program. Do NOT open in Word or a PDF program. Scroll past the barcode sequences to find the analysis script.
- The script is a loop that counts the number of reads aligned to each PTM-specific DNA barcode in a reaction. Each PTM in the SNAP-CUTANA Panel is represented by two unique barcodes, A & B. For the script, you need to create one loop per control reaction. To customize:
a. Copy lines between # template loop begin ## and # template loop end ##.
b. Paste the loop under the last done. Paste one copy per control reaction.
c. In the first loop replace sample1_R1.fastq and sample1_R2.fastq with R1 & R2 fastq file names for one control reaction. Repeat for each loop. Press save. - In Terminal, set the directory to your folder: Type cd and press space. Drag the folder from your files into Terminal to copy the location. Press return.
- Run your script in Terminal: Type sh and press space. Drag your .sh file from your files into Terminal to copy the file location. Press return. Terminal generates barcode read counts from R1 & R2 reads, one loop/reaction at a time.
- Open the Panel Analysis Excel .xlsx file. Fill in reaction names and set the on-target PTM in Column B. The first reaction is set to IgG (negative control); for other reactions, select a target (i.e. H3K4me3) from the drop-down menu.
- Copy R1 barcode read counts from the first loop in Terminal. In Excel, paste into the yellow cells for that reaction in Column C. Copy & paste the R2 read counts from the same loop to yellow cells in Column D. Repeat for each loop/reaction.
- The Excel file automatically analyzes spike-in data for each reaction by:
a. Calculating total read counts for each DNA barcode (R1 + R2) in Column E.
b. Calculating total barcode read counts for each PTM (A + B) in Column F.
c. Expressing total read counts for each PTM as a percentage of on-target PTM read counts (Columns G & J), providing a readout of on- vs. off-target PTM recovery and antibody specificity. - Column J auto-populates the Output Table (Figure). Reactions are separated by row and PTM data are sorted into columns. A color gradient is used to visualize the recovery of each PTM normalized to on-target PTM, from blue (100%) to orange (less than 20%).
- For each reaction, calculate the percent of unique sequencing reads that have been assigned to spike-ins. In Excel, type the total number of unique reads in the yellow cell Uniq align reads (in Column B). The % total barcode reads is calculated in the cell immediately below and is added to the Output Table.
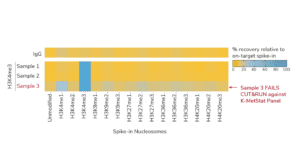
Figure. K-MetStat Spike-ins validate workflows and flag poor samples in CUTANA CUT&RUN experiments. Spike-in data for H3K4me3 positive control reactions is shown for three independently prepared mouse B cell samples (10,000 cells each). Samples 1 & 2 show expected results, while Sample 3 was flagged for recovery of off-target PTMs and low signal-to-noise. Representative data from one IgG reaction is shown as a negative control.
Expected results from SNAP-CUTANA Spike-in control reactions
- The IgG negative control shows low background and no preference among PTMs (Figure, Top row).
- H3K4me3 positive control has strong enrichment for H3K4me3 spike-ins, less than 20% off-target PTM recovery, and high signal-to-noise (Figure, Samples 1 and 2).
- Spike-in barcode reads comprise ~1% (0.5-5%) of total sequencing reads. This may vary based on target abundance and sequencing depth. The main goal is to have thousands of reads, which will allow adequate sampling of the K-MetStat Panel for reliable data analysis.
- If control reactions generate expected spike-in data (Figure, Samples 1 and 2), you can be confident in the technical aspects of your workflow.
- More than 20% off-target PTM recovery in H3K4me3 control and/or high background in IgG control indicate experimental problems (Figure, Sample 3). See this article for guidance for using SNAP-CUTANA Spike-in controls to troubleshoot workflows.
SNAP-CUTANA Spike-in Controls
Guidance on our nucleosome controls
What are SNAP-CUTANA™ Spike-ins?
SNAP-CUTANA Spike-in Controls are a defined spike-in control added to cell samples at the start of CUT&RUN experiments. They are specifically designed for reactions targeting histone post translational modifications (PTMs), such as histone lysine methylation, and allow users to examine histone PTM antibody performance, workflow success, and much more. Because SNAP-CUTANA Spike-ins replicate chromatin structure (i.e. nucleosomes), the natural target of CUT&RUN, they provide accurate on- and off-target substrates for histone PTM antibodies.
Features and advantages include:
- Fast: SNAP-CUTANA Spike-ins are coupled to magnetic beads, allowing them to be added to CUT&RUN reactions in one quick step for seamless workflow integration (Figure 1).
- Easy: No protocol modifications are necessary. SNAP-CUTANA Spike-in data are quickly analyzed in final sequencing data using our step-by-step instructions.
- Reliable: Use these controls to examine sample quality, MNase activity, antibody specificity, and troubleshoot challenging workflows.

Figure 1. SNAP-CUTANA Spike-in Controls are pools of highly pure nucleosomes carrying defined histone PTMs. For instance, the SNAP-CUTANA K-MetStat Panel shown in this figure is a pool of 16 nucleosomes representing 16 distinct methyl-lysine states. Spike-in nucleosomes are individually coupled to magnetic beads and pooled into a single panel for convenient one-step addition to CUT&RUN workflows. pAG-MNase cleavage releases all antibody-bound targets into solution, including antibody-bound spike-ins. Each spike-in nucleosome contains a PTM-specific DNA barcode sequence, which enables detection of K-MetStat Panel controls in sequencing data.
How do SNAP-CUTANA Spike-ins work in CUT&RUN?
You can add SNAP-CUTANA Spike-in Controls to reactions that are mapping a histone PTM included in the Panel. SNAP-CUTANA Spike-ins are added to reactions just prior to addition of target-specific antibody, in Section IV of the CUT&RUN Protocol (Figure 2).
Immobilization of spike-in nucleosomes on magnetic beads makes them similar to bead-coupled cells, allowing both to be captured using magnetic separation racks. This means that spike-ins can be added early in the experiment for side-by-side processing with bead-coupled cells, all in the same reaction tube. Spike-ins can thus report on multiple aspects of the CUT&RUN workflow, including antibody specificity and pAG-MNase activity.
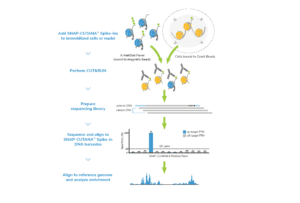
Figure 2. Schematic showing how SNAP-CUTANA Spike-ins, such as the K-MetStat Panel, are used during CUT&RUN workflows.
No other modifications to the protocol are required. You simply continue the CUT&RUN workflow, adding your target specific antibody for overnight incubation. Antibodies bind their specific histone PTM target in permeabilized cells AND in the SNAP-CUTANA Spike-in Panel. The next day, pAG-MNase is used to cleave antibody-bound chromatin (in cells) and antibody-bound spike-ins.
Cleaved spike-ins are released from magnetic beads into solution along with the clipped DNA fragments from cells. A magnet is used to removed unclipped spike-ins and cells, and target fragments are purified from the supernatant for sequencing.
The PTM-specific DNA barcode sequences on the spike-in nucleosomes (Figures 1 and 2) are used to detect reads from the spike-ins vs. cells in sequencing data. Generally, we aim for ~1% of total sequencing reads to be from SNAP-CUTANA Spike-ins, although this may vary based on sequencing depth and target abundance. The main goal is to have many thousands of sequencing reads aligned to SNAP-CUTANA Spike-ins, which enables adequate sampling of the panel and reliable use in downstream applications.
When assessing the reads assigned to spike-ins, you should see enrichment for on-target PTM and minimal reads from off-target PTMs in the Panel (Figure 2). These data can be used as a direct readout for PTM recovery and assay success, described in more detail here.
How to analyze SNAP-CUTANA™ Spike-in Controls
- Download R1 & R2 paired-end sequencing files (fastq.gz) for control reactions. Double-click the fastq.gz files to create fastq files and save in a new folder.
- On the SNAP-CUTANA Spike-in product page (e.g. K-MetStat Panel), under Documents and Resources, download the Shell Script (.sh) and K-MetStat Panel Analysis (.xlsx) files. Save to the folder from Step 1.
- Open the .sh file in TextEdit or any text editing program. Do NOT open in Word or a PDF program. Scroll past the barcode sequences to find the analysis script.
- The script is a loop that counts the number of reads aligned to each PTM-specific DNA barcode in a reaction. Each PTM in the SNAP-CUTANA Panel is represented by two unique barcodes, A & B. For the script, you need to create one loop per control reaction. To customize:
a. Copy lines between # template loop begin ## and # template loop end ##.
b. Paste the loop under the last done. Paste one copy per control reaction.
c. In the first loop replace sample1_R1.fastq and sample1_R2.fastq with R1 & R2 fastq file names for one control reaction. Repeat for each loop. Press save. - In Terminal, set the directory to your folder: Type cd and press space. Drag the folder from your files into Terminal to copy the location. Press return.
- Run your script in Terminal: Type sh and press space. Drag your .sh file from your files into Terminal to copy the file location. Press return. Terminal generates barcode read counts from R1 & R2 reads, one loop/reaction at a time.
- Open the Panel Analysis Excel .xlsx file. Fill in reaction names and set the on-target PTM in Column B. The first reaction is set to IgG (negative control); for other reactions, select a target (i.e. H3K4me3) from the drop-down menu.
- Copy R1 barcode read counts from the first loop in Terminal. In Excel, paste into the yellow cells for that reaction in Column C. Copy & paste the R2 read counts from the same loop to yellow cells in Column D. Repeat for each loop/reaction.
- The Excel file automatically analyzes spike-in data for each reaction by:
a.Calculating total read counts for each DNA barcode (R1 + R2) in Column E.
b. Calculating total barcode read counts for each PTM (A + B) in Column F.
c. Expressing total read counts for each PTM as a percentage of on-target PTM read counts (Columns G & J), providing a readout of on- vs. off-target PTM recovery and antibody specificity. - Column J auto-populates the Output Table (Figure). Reactions are separated by row and PTM data are sorted into columns. A color gradient is used to visualize the recovery of each PTM normalized to on-target PTM, from blue (100%) to orange (less than 20%).
- For each reaction, calculate the percent of unique sequencing reads that have been assigned to spike-ins. In Excel, type the total number of unique reads in the yellow cell Uniq align reads (in Column B). The % total barcode reads is calculated in the cell immediately below and is added to the Output Table.

Figure. K-MetStat Spike-ins validate workflows and flag poor samples in CUTANA CUT&RUN experiments. Spike-in data for H3K4me3 positive control reactions is shown for three independently prepared mouse B cell samples (10,000 cells each). Samples 1 & 2 show expected results, while Sample 3 was flagged for recovery of off-target PTMs and low signal-to-noise. Representative data from one IgG reaction is shown as a negative control.
Expected results from SNAP-CUTANA Spike-in control reactions
- The IgG negative control shows low background and no preference among PTMs (Figure, Top row).
- H3K4me3 positive control has strong enrichment for H3K4me3 spike-ins, less than 20% off-target PTM recovery, and high signal-to-noise (Figure, Samples 1 and 2).
- Spike-in barcode reads comprise ~1% (0.5-5%) of total sequencing reads. This may vary based on target abundance and sequencing depth. The main goal is to have thousands of reads, which will allow adequate sampling of the K-MetStat Panel for reliable data analysis.
- If control reactions generate expected spike-in data (Figure, Samples 1 and 2), you can be confident in the technical aspects of your workflow.
- More than 20% off-target PTM recovery in H3K4me3 control and/or high background in IgG control indicate experimental problems (Figure, Sample 3). See this article for guidance for using SNAP-CUTANA Spike-in controls to troubleshoot workflows.
In the CUTANA™ CUT&RUN Kit, EpiCypher adds the SNAP-CUTANA™ K-MetStat Panel to positive (H3K4me3) and negative (IgG) control reactions to confirm workflows and guide troubleshooting. Each CUT&RUN kit comes with sufficient controls antibodies and the K-MetStat Panel sufficient for 10 experiments:
- 10 reaction volumes of IgG Negative Control Antibody
- 10 reaction volumes of H3K4me3 Positive Control Antibody
- 20 reaction volumes of the SNAP-CUTANA K-MetStat Panel (added to both positive and negative control reactions)
Of note, EpiCypher often uses H3K27me3 as an additional positive control (EpiCypher 13-0055), as it has higher yields than H3K4me3. The K-MetStat Panel should also be added to H3K27me3 control reactions; purchase additional K-MetStat Panel here.
Why are these controls useful?
SNAP-CUTANA Spike-in controls are the only control that replicates the in vivo target of CUT&RUN (i.e. nucleosomes). The spike-ins are added to cells just after ConA bead binding and are processed alongside the sample throughout the CUT&RUN protocol (Figure 1). By combining these spike-ins with our rigorously validated CUT&RUN antibodies for H3K4me3 and IgG, we can immediately gauge workflow success.
EpiCypher includes these control reactions in every experiment, and it has saved us countless time and resources in protocol optimization. Should you reach out to our technical support team for troubleshooting assistance, they will be able to assist you much better if you include data derived from this panel. For details on how to leverage the K-MetStat Panel for troubleshooting, see this article.

Figure 1. The K-MetStat Panel is composed of 16 spike-in nucleosomes, representing 15 distinct histone lysine methylation PTMs and an unmodified control. Nucleosomes are coupled to magnetic beads for easy one-step addition to CUT&RUN. A PTM-specific DNA barcode enables detection of spike-ins in sequencing data.
Pairing the K-MetStat Panel with control antibodies to determine assay success
We review the specifics of how SNAP-CUTANA Spike-ins are processed in CUT&RUN reactions here; the strategy using the K-MetStat Panel with control antibodies is outlined in Figure 2. Briefly:
- Add the SNAP-CUTANA K-MetStat Panel to designated control reactions immediately prior to the addition of H3K4me3 or IgG control antibody (Figure 2).
- Add antibody, which binds its target in cells and in the spike-in panel (Figure 2).
- pAG-MNase cleaves antibody-bound chromatin and antibody-bound spike-in. Cleaved DNA is purified and prepared for sequencing.
- Samples are sequenced. For each control reaction, download sequencing files and determine the number of sequencing reads aligned to each PTM-specific DNA barcode, using the analysis procedure as described in this article. Barcode read counts provide a useful measurement of PTM recovery and workflow success.

Figure 2. The K-MetStat Panel is used with control antibodies to determine CUT&RUN success. Two different outcomes are shown. Note that the K-MetStat Panel can be added to experimental reactions targeting a PTM in the Panel. Additional K-MetStat Panel can be purchased at here.
What are the expected results? How is assay success determined?
Figure 2 illustrates two potential outcomes from H3K4me3 and IgG control reactions: one successful CUT&RUN experiment and one that indicates a failed experiment. In each heatmap, reactions are separated into rows and recovery of each PTM in the Panel is separated by column. The heatmap shows data normalized to the on-target PTM (i.e. the on-target PTM is set to 100%, appearing blue). Off-target PTMs should have low signal relative to this target and appear orange. EpiCypher always aims for off-target PTMs to show <20% signal relative to the target PTM.
To determine successful CUT&RUN, keep the following metrics in mind:
- The IgG negative control should show no preference among PTMs and low background.
- H3K4me3 positive control should have strong enrichment for H3K4me3 spike-ins, less than 20% off-target PTM recovery, and high signal-to-noise.
- Spike-in barcode reads should comprise ~1% (0.5-5%) of total sequencing reads. This may vary based on target abundance and sequencing depth. The main goal is to have thousands of reads, which will allow adequate sampling of the K-MetStat Panel for reliable data analysis.
- If control reactions generate expected spike-in data, you can be confident in the technical aspects of your workflow (Figure 2, left heatmap).
- If more than 20% off-target PTM recovery in H3K4me3 control and/or high background in IgG control indicate experimental problems (Figure 2, right heatmap). See this article for a discussion of troubleshooting using spike-in results.
It may be unclear from genomic tracks alone if a reaction issue has occurred. Pairing SNAP-CUTANA™ Spike-in Controls with EpiCypher’s robust control antibodies enables users to flag failed reactions AND identify potential causes for troubleshooting. Together, these controls allow researchers to be confident in their experimental results.
Here, we will discuss how to use SNAP-CUTANA Spike-in results to troubleshoot problematic workflows. To learn how to apply SNAP-CUTANA Spike-ins to confirm workflow success, see this article.
Figures 1 and 2 demonstrate the use of K-MetStat Spike-in data for troubleshooting.
- In Figure 1 we used spike-in data from H3K4me3 & IgG control reactions to validate workflows for three independently prepared mouse B cell samples.
- Samples 1 & 2 showed expected results from control reactions, while Sample 3 displayed low signal-to-noise (S:N) and high off-target PTM recovery (Figure 1).
- Genomic profiles agreed with spike-ins: Samples 1 & 2 generated expected tracks for H3K4me3 & H3K27me3, while Sample 3 profiles had poor S:N (Figure 2).

Figure 1. K-MetStat Spike-ins validate workflows and flag poor samples in CUTANA CUT&RUN experiments. Spike-in data for H3K4me3 positive control reactions is shown for three independently prepared mouse B cell samples (10,000 cells each; protocol optimization experiment with a multi-lab consortium). Samples 1 & 2 show expected results, while Sample 3 was flagged for recovery of off-target PTMs and low signal-to-noise. Representative data from one IgG reaction is shown as a negative control.
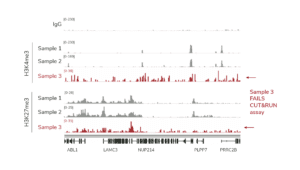
Figure 2. Genomic tracks featuring data in Figure 1. CUT&RUN was used to map IgG (negative control), H3K4me3 (positive control) and H3K27me3 in three independently prepared mouse B cell samples (10,000 cells each; protocol optimization experiment with a multi-lab consortium). A representative 400 kb region is shown. Samples 1 and 2 show consistent peaks, while Sample 3 displays low S:N (red).
To troubleshoot Sample 3 reactions, we considered the following:
- All reactions were performed in parallel using the same antibodies and reagents. However, only Sample 3 reactions had problems with background.
- Sample 3 showed poor S:N in both genomic profiles and K-MetStat Panel data.
- Sample 3 generated poor profiles across multiple targets.
Combined, these results suggested problems with sample prep vs. a complete workflow failure. We subsequently reviewed Sample 3 processing methods, revealing that the number of cells used per reaction was much lower than intended. For other troubleshooting tactics using the K-MetStat Panel, see Table 1 (below).
Table 1. Troubleshooting using SNAP-CUTANA Spike-ins
| Results | Causes and troubleshooting approaches |
| K-MetStat spike-in data:
• High target specificity • High S:N • Genomic data: Poor S:N |
pAG-MNase cleavage and wash conditions are optimized. Control antibodies are performing as expected. Problems may include:
⚠ Low numbers of cells • Optimize assay with 500,000 cells before decreasing input • If using nuclei, adherent cells, cross-linked cells, tissues, or cryopreserved samples, see this section for modifications ⚠ Poor sample prep • Optimize Digitonin permeabilization of cells • Confirm sample integrity and bead binding • Avoid ConA bead clumping and dry out during assay ⚠ Experimental target requires different processing conditions • Ensure target is present and localized to chromatin • If using frozen cells, try freshly isolated cells • Test native vs. lightly cross-linked conditions |
| K-MetStat spike-in data:
• Nonspecific PTM recovery • Poor S:N • Genomic data: Poor S:N |
⚠ Indicates a fundamental failure in the workflow
• Carefully re-read the protocol and important notes • Ensure buffers are prepared fresh on day of use • Ensure ConA beads are in good condition (e.g. never frozen) • Make sure correct parameters are used in indexing PCR; consider using the CUTANA CUT&RUN Library Prep Kit ⚠ Low numbers of cells and/or poor sample prep • Optimize following the guidelines above |
| K-MetStat spike-in data:
• Nonspecific PTM recovery • S:N may vary • Genomic data: High S:N |
⚠ Indicates cross-reactive control antibodies
• Examine potential contamination of control reactions with antibodies to other targets • Ensure buffers are prepared fresh on day of use • Change pipette tips after each reagent addition to avoid cross-contamination • For concerns about control antibody performance, email us at technical@stratech.co.uk |
At EpiCypher, we have developed two main applications of the SNAP-CUTANA Spike-in Controls for CUT&RUN. The first is determination of workflow success, discussed here. The other application of SNAP-CUTANA Spike-ins is for histone PTM antibody validation.
Why is testing histone PTM antibodies important?
Histone PTM antibodies are notoriously nonspecific, which can result in misleading biological interpretations. EpiCypher is well aware of this problem. We first developed SNAP nucleosome spike-in technology for ChIP-seq (SNAP-ChIP Spike-ins) and used them to test more than 400 lysine-methylation and lysine-acylation antibodies in ChIP assays. We found that >70% of histone PTM antibodies show significant off-target binding and/or low binding efficiency – even highly cited antibodies! The results of this study can be found at chromatinantibodies.com.
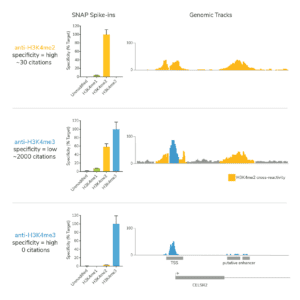
Figure 1. A highly cited H3K4me3 antibody fails SNAP Spike-in testing and shows extensive cross-reactivity to H3K4me2 (middle panel). Highly specific H3K4me2 (top panel; orange) and H3K4me3 (bottom panel; blue) antibodies are shown for comparison. See Shah et al. 2018.
These results were a major driving factor in the development of SNAP-CUTANA Spike-in technology. To ensure the success of emerging CUT&RUN (and CUT&Tag) epigenomic assays, defined controls are needed to find the best antibodies possible.
Why should I use SNAP-CUTANA Spike-ins to validate antibodies, compared to other methods?
This strategy is the only method that directly confirms antibody specificity in CUT&RUN against physiological on- and off-target substrates. EpiCypher knows from experience that histone PTM antibodies do not perform the same across assays – for instance, a good ChIP antibody is NOT guaranteed to work in CUT&RUN!
EpiCypher metrics for SNAP-CUTANA CUT&RUN antibody validation:
EpiCypher is using these defined nucleosome spike-in controls to identify best-in-class histone PTM antibodies for CUT&RUN. We offer a collection of SNAP-Certified Antibodies for CUT&RUN and can provide antibody recommendations upon request.
We validate antibody specificity as well as efficiency, allowing users to be confident when using reduced cell numbers. Each of our SNAP-Certified™ Antibodies show:
- High specificity: <20% recovery of off-target PTMs
- High target efficiency: Robust profiling at 500,000 and 50,000 starting cells
To use SNAP-CUTANA Spike-ins for PTM antibody testing:
- Purchase a SNAP-CUTANA Panel that contains your histone PTM target. For instance, you can use the K-MetStat Panel for any reaction that is mapping a histone methyl-lysine mark included in the panel.
- Source several antibodies to your histone PTM target to test, ideally from several sources and/or targeting distinct epitopes.
- Test antibodies side-by-side in CUT&RUN using 500,000 native cells per reaction.
- Add the SNAP-CUTANA Spike-in Panel to CUT&RUN reactions just before addition of primary antibody, as indicated in the protocol. Include control reactions with the K-MetStat Panel to confirm workflow success.
- Aim for <20% antibody cross-reactivity and consistent genomic enrichment with 500,000 cells. To help ensure high antibody efficiency, validation at 50,000 cells is also recommended.
Several notes:
- Do NOT add a SNAP-CUTANA Panel to reactions targeting a protein or a PTM that is not represented in the panel.
- Note that for CUTANA CUT&RUN Kit users, this will require purchase of additional K-MetStat Panel.
Troubleshooting and FAQs
The most commonly asked questions
There are many steps to consider when troubleshooting CUT&RUN assays. A good place to start is by reviewing the basic CUT&RUN troubleshooting guidelines below. Alternatively, navigate to the specific topics.
- Low CUT&RUN yields (before library prep)
- Fragment distribution and library yields (after library prep)
- CUT&RUN sequencing and results
Basic CUT&RUN troubleshooting guidelines:
See the quality control metrics in the Figure below. If your experiment failed or if you have very low yields (< 5ng), see this article and consider the following questions:
- What is your cell/sample type? Review sample prep protocol modifications.
- Are Digitonin permeabilization conditions optimized for your cell type?
- Have you confirmed sample prep and ConA bead binding?
- Are you using the recommended 500,000 cells per reaction? Success from low cell numbers depends on antibody quality and target abundance. Review this article for guidance.
- Have you included reactions with control antibodies & the K-MetStat Panel? These controls are crucial for troubleshooting CUT&RUN.
- Confirm proper ConA bead storage and condition. Are ConA beads brown and stored at 4˚C? ConA beads should NEVER be frozen.
- Have reactions been mixed properly using a nutator? Have ConA beads become clumpy or dried out during the protocol?

Figure. Overview of CUTANA™ CUT&RUN kit quality ensure metrics and checks to ensure successful chromatin profiling. Quality control metrics are listed for each section of the CUT&RUN workflow.
Troubleshooting fragment distribution and library yields (after library prep)
| Concern | Potential causes & troubleshooting approach |
| Low library yields, no enrichment in Bioanalyzer or TapeStation results | ⚠ Low CUT&RUN yields, low inputs for library prep
• See this article for guidance ⚠ Library prep technique • Use the EpiCypher CUTANA CUT&RUN Library Prep Kit (EpiCypher 14-1001 & 14-1002), specifically developed for CUT&RUN |
| Adapter dimers | ⚠ Self-ligation of sequencing adapters, preferentially amplified due to their small size (see this FAQ)
• Keep adapter ligation reagents on ice during ligation setup • Remove adapter dimers comprising >5% of a library; see the CUT&RUN Library Prep Kit Manual |
Troubleshooting CUT&RUN sequencing and results
| Concern | Potential causes & troubleshooting approach |
| Sequencing a low-concentration DNA library | ⚠ If it is not possible to repeat library prep:
• Use a Speedvac to increase the library concentration • Add as much of the library as possible to the sequencing pool • Deeper sequencing is recommended |
| Background in open chromatin | ⚠ Indicates over-digestion by MNase
• Repeat assay with fresh buffers • Make sure MNase digestion is performed on ice |
| Experimental target shows high background and/or is indistinguishable from IgG negative control | ⚠ Over-digestion by MNase, DNA damage, antibody failure. Use 500,000 cells per reaction; include reactions with control antibodies & the K-MetStat Panel. General tips:
• Process cells quickly and resuspend in cold Antibody Buffer • Test multiple antibodies to experimental target • Ensure MNase digestion is incubated on ice for 2 hours • Keep adapter ligation reagents on ice during ligation setup |
Use an antibody validated in CUT&RUN for best chance of success.
For your convenience, EpiCypher offers an array of CUT&RUN certified antibodies for histone post-translational modifications (PTMs) and chromatin associated proteins, all validated in our CUTANA™ CUT&RUN assays. When beginning a CUT&RUN experiment, we recommend first searching our catalog of CUT&RUN validated antibodies.
Don’t see your target? See our articles about CUT&RUN antibody validation for histone PTM or chromatin associated proteins for CUT&RUN, or contact us for recommendations.
You may try, but fair warning: EpiCYpher has found that success in ChIP does NOT guarantee success in CUT&RUN.
This is largely due to differences in sample prep and processing steps. ChIP uses heavily cross-linked cells, stringent wash buffers, and bead-coupled antibodies to help maximize the signal-to-noise ratio. However, these strategies often lead to loss of on-target signal, which is particularly problematic for low abundance targets. To counteract these effect, ChIP requires highly efficient antibodies, with high yields.
In contrast, CUT&RUN uses native chromatin, mild washes, and antibodies in solution, reflecting the increased sensitivity of this newer technique. The only way to know if your antibody will work in CUT&RUN if the antibody has been tested in this assay – antibody validation using ChIP, immunoblot, ELISA, IHC, or other techniques is NOT a predictor of CUT&RUN performance.
For your convenience, EpiCypher offers CUT&RUN-validated antibodies, which you can shop here.
As outlined in this article, EpiCypher checks sample integrity at three steps during the CUT&RUN protocol. It is crucial that samples pass all three sample checks, as poor quality samples increase assay background and reduce signal-to-noise.
EpiCypher focuses on several features when it comes to assessing cell quality: morphology/integrity, viability, and total cell counts. Ask yourself these key questions when analyzing sample quality:
What is the normal morphology for my cell type?
EpiCypher routinely works with K562 cells, which have rounded cell morphology. It is key that the cells appear normal before attaching to ConA beads.
What is the optimal viability for my cell type?
For freshly harvested, native K562 cells, EpiCypher typically observes >90% viability using Trypan Blue staining. However, viability may vary drastically by cell type or experimental conditions, such as drug treatments. Furthermore, some cells display reduced viability in CUT&RUN Wash Buffer. Be sure to follow our instructions for assessing sample quality and make the best decision based on your sample type.
Do I have the appropriate number of cells?
EpiCypher recommends using 500,000 cells per CUT&RUN reaction. Count cells both at time of harvest and before ConA bead binding. The second count will confirm you have not lost significant sample during prep and ensure confirm cell morphology before proceeding with the experiment.
Are my cells being accurately counted and assessed?
Trypan Blue is toxic to cells, and some cells may display high sensitivity. As such, it is important that Trypan Blue dye is added immediately before cell counting. If your cells show low viability, yet have normal morphology, minimal debris/lysis, and have been successfully expanded in culture, it may be prudent to test a different cell counting method. Propidium Iodide is a similar nuclear dye that has been implemented with success in several collaborator labs. Alternatively, Trypan Blue can be used at a lower dilution.
It is normal to lose viability following freeze-thaw, as cells may be more permeable to Trypan Blue. For frozen samples, EpiCypher scientists often focus more on cellular morphology. Total cell counts are important, as is the balance between good cell integrity vs. cell lysis. As long as you have not lost a substantial portion of cells, and there is minimal cell lysis, you should be fine to move forward with CUT&RUN.
For nuclei preparations, >95% nuclei should be Trypan Blue positive or “dead” and unclumped. Nuclei should be round, intact, and contain minimal debris (as this could indicate lysis; see Figure 1). Use Trypan Blue staining to monitor and optimize nuclear extraction, increasing spin time if losing sample.
Note: to best ensure success, we suggest using our nuclei prep protocol.

Figure 1. Morphology characteristic of intact K562 cells (left) compared to isolated nuclei (right) when visualized under brightfield microscope after Trypan Blue staining. Isolated nuclei will stain blue, while cells will be bright white and round. For accurate nuclei counts, record “dead” cell numbers on an automated cell counter or manually count blue stained nuclei.
CUT&RUN success depends on many factors, including cell type, cell number, target abundance, and antibody quality. The Figure below outlines optimization guidelines.
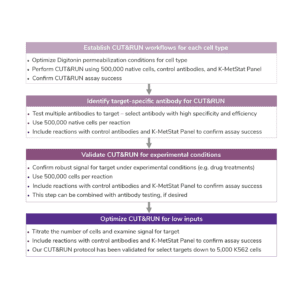
Figure. Development and optimization guides for successful CUT&RUN workflows.
See the Figure below for optimization guidelines. Note that using low cell numbers may result in lower signal and increased background, including for control antibodies.
Lower yields may be partly due to antibody quality and target abundance, as well as reduced cell numbers. Furthermore, an antibody that works well with 500,000 cells may fail at lower inputs.
Library prep can be optimized for low yields. Deeper sequencing is often recommended.

Figure. Development and optimization guidelines for successful CUT&RUN experiments.
CUTANA™ CUT&RUN workflows contain multiple quality control checks to help ensure assay success. This includes the control reactions and spike-in controls outlined below, as well as key cell sample quality checks, confirmation of cell binding to ConA beads, examination of CUT&RUN DNA yields, and fragment distribution analysis of purified sequencing libraries. Find a summary of CUTANA CUT&RUN quality control steps here.
Control antibodies
Reactions using negative control (IgG) and positive control (H3K4me3) antibodies should be included in every experiment to validate protocols and examine assay background. When used with SNAP-CUTANA™ Spike-in controls (below), control antibodies can assist in validating and troubleshooting your workflow.
Spike-in nucleosome controls
SNAP-CUTANA™ Spike-in Controls are the only control that uses purified recombinant nucleosomes, replicating the physiological target of CUT&RUN assays.
SNAP-CUTANA Spike-in Controls are essential controls for any CUT&RUN experiment. When combined with negative and positive control antibodies, SNAP-CUTANA Spike-in Controls give users the best chance of CUT&RUN success. For more information on SNAP-CUTANA Spike-in Controls, visit their section or navigate to specific articles on:
- What are SNAP-CUTANA Spike-in Controls and how do they work?
- Analyzing SNAP-CUTANA Spike-in Controls
- Using SNAP-CUTANA Spike-in Controls to determine CUT&RUN success
- Using SNAP-CUTANA Spike-in Controls to troubleshoot workflows or validate select
- Using SNAP-CUTANA Spike-in Controls to troubleshoot workflows or validate select histone post-translational modification antibodies
E. coli Spike-in DNA
E. coli DNA is spiked in prior to library prep to aid in library prep and sequencing troubleshooting as well as normalization. For more information on how to use E. coli spike-in DNA for normalization, see this article.
SNAP-CUTANA™ Spike-ins are the only control that can be used for CUT&RUN optimization, workflow monitoring, antibody validation, and troubleshooting. EpiCypher recommends pairing the SNAP-CUTANA K-MetStat Panel with our validated H3K4me3 positive and IgG negative control antibodies in each experiment to help ensure experimental success.
The K-MetStat Panel is a defined spike-in control
Although the SNAP-CUTANA K-MetStat Panel works like other genomic spike-ins, its design comes with added benefits. The K-MetStat Panel is the only control that uses purified recombinant nucleosomes, replicating the physiological target of CUT&RUN assays. The panel comprises 16 unique nucleosomes, each containing a distinct histone lysine methylation PTM (or unmodified) and DNA barcode, which provides both on- and off-target substrates for control reactions.
Streamlined control for improved CUT&RUN
The K-MetStat Panel is directly added to cells and processed alongside the sample throughout the CUT&RUN protocol in one easy step (see Figure 1 below). The spike-in data can be used to validate specific aspects of the workflow, such as antibody specificity or enzyme activity, and provide key insights for troubleshooting. For information on how to use the K-MetStat Panel for workflow validation and monitoring technical variability, see this article.
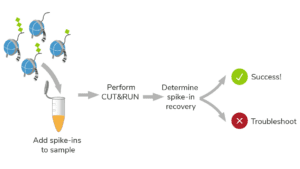
Figure 1. SNAP-CUTANA Spike-in Controls are processed alongside samples for a direct, in-assay readout of experimental success.
ConA beads are crucial to the CUT&RUN workflow. They should be stored at 4˚C, appear brown in color, and be easily mixed by gentle vortexing or pipetting.
- Do NOT use ConA beads that have been frozen – this damages beads.
- Do NOT use ConA beads that appear black, granular, or clumpy.
- Activated ConA beads should be kept on ice and used within four hours of activation.
After ConA bead binding, save an aliquot of supernatant [unbound fraction] and take an aliquot of sample slurry [bead fraction] for Trypan blue staining as described. Successful ConA bead binding is indicated when:
- The unbound fraction contains few cells/nuclei (Figure A).
- The bead fraction contains permeabilized, Trypan Blue positive cells surrounded by beads (Figure B).
Should you not observe this, ensure:
- Count/integrity of cells before binding to ConA beads.
- ConA beads were never frozen.
- Cells/nuclei were not clumped.
- Beads did not become clumped or dried out.
- All buffers were correctly prepared.

Figure. Unbound fraction from ConA bead prep shows few cells/nuclei (A), whereas the bead fraction contains permeabilized (Trypan Blue positive) cells/nuclei surrounded by beads (brown spheres).
ConA beads typically become clumpy/sticky after overnight incubation at 4˚C. Some clumping is normal, and should not impact your data, especially if you are starting with 500,000 high-quality cells. Beads can be dispersed by gentle pipetting. The end of a pipette tip can be cut off to help mix or preserve delicate cells.
However, if you start with a sample of poor quality – meaning lysed cells and/or poor cellular integrity – you may experience more ConA bead clumping.
Excessive bead clumping leads to sample loss, reduces yields, and negatively impacts quality. Thus, it is key to minimize bead clumping as much as possible during your CUT&RUN experiment. During pAG-MNase binding and digestion, it is particularly important that ConA beads are evenly resuspended.
There are several steps you can take to help reduce ConA bead clumping and precipitation.
- Do NOT let ConA beads dry out. Avoid disturbing beads with pipette while on magnet.
- Do NOT rotate or invert tubes. Rotation causes ConA beads to stick to tube sides and dry out, reducing yields. Use a nutator for incubations and elevate tube caps.
Note: if accidental inversion occurs, quick spine tubes immediately to collect material. - Be careful when pipetting very sticky beads, as they can stick to the inside of pipette tips and cause sample loss. In cases with excessive bead clumping, gently vortexing is preferred.
- Start with high-quality cells, confirmed using our quality control checks. Increased cell lysis can cause ConA beads to become more clumpy, reducing yields.
TapeStation/Bioanalyzer and qPCR will NOT provide useful information at this step of the workflow. CUT&RUN yields are too low for these methods. Read on for more information as to why.
Why can’t I use qPCR?
qPCR is used in ChIP to verify the enrichment of a known on-target region compared to the bulk chromatin input. However, CUT&RUN is performed in intact cells. There is no immunoprecipitation step and no bulk chromatin input for comparison. Simply stated, there is no way to properly control or analyze qPCR.
Why can’t I use the Bioanalyzer/TapeStation?
CUT&RUN uses intact cells bound to a solid support and selectively cleaves antibody-bound chromatin. These advances bypass bulk chromatin fragmentation and immunoprecipitation, resulting in high signal-to-noise and low cell input requirements vs ChIP. As a result, however, raw CUT&RUN DNA yields are often below the limit of sensitivity for fragment size distribution using the TapeStation or Bioanalyzer.
Low CUT&RUN DNA yields are common for low abundance targets (e.g. H3K4me3), but also depend on the number/quality of starting cells, cell type, and antibody performance. If you are consistently generating low yields for experimental targets:
- Review Basic CUT&RUN troubleshooting guidelines and be sure to always include all QC checks outlined in the Figure below. Pay careful attention to the quality and count of your cells during sample prep and avoid ConA bead dry out, which causes sample loss.
- Include H3K4me3 and IgG control reactions. If these controls work but experimental targets fail, confirm that your target is correctly localized to chromatin (e.g. stimulation conditions) and test additional antibodies and/or cross-linking conditions.
- If the experiment cannot be repeated, use the total amount of CUT&RUN-enriched DNA for library prep. See the CUTANA™ CUT&RUN Library Prep Kit manual for guidance.
NOTE: For some targets and cell types, low CUT&RUN yields are unavoidable. For guidance on these situations, see this article.

Figure. Quality control metrics for each step of the CUTANA CUT&RUN kit.
Similar yields from H3K4me3 and IgG controls do NOT imply CUT&RUN failure. H3K4me3 is a low abundance target, resulting in lower yields that are often similar to IgG. In these cases, use the total amount of CUT&RUN-enriched DNA for library prep. In EpiCypher’s experience, good sequencing data with high signal-to-noise are still obtained.
If desired, a high abundance target (e.g. H3K27me3, EpiCypher 13-0055) can be used as an additional positive control, as yields will be much higher than IgG.
Ideally, you would troubleshoot low CUT&RUN yields, repeat the experiment, and then perform library prep and sequence. This will result in higher quality sequencing data compared to using ultra-low CUT&RUN yields. However, in some cases, repeating the experiment isn’t possible. In fact, for some targets and cell types, low CUT&RUN yields are unavoidable.
Although useful sequencing data can be obtained, the resulting libraries often have low concentrations with elevated adapter dimers, reduced read diversity, and low signal over background, all of which impact data quality.
Use the CUTANA CUT&RUN Library Prep Kit (EpiCypher 14-1001 & 14-1002), which is specifically optimized for CUT&RUN workflows and includes guidelines for library prep from low CUT&RUN yields (see manual).
General tips:
- Remove adapter dimers that have greater than 5% “%Integrated area” (as called by TapeStation analysis), meaning that the adapter dimers comprise >5% of your library. High levels of adapter dimers take up valuable sequencing bandwidth. See our articles on avoiding or removing adapter dimers for more.
- Increase number of cycles for indexing PCR to improve library yields for Bioanalyzer/TapeStation analysis and sequencing. Refer to the CUTANA CUT&RUN Library Prep Kit manual for guidance.
- Deeper sequencing (>8 million reads) is recommended to capture read diversity.
- Read duplicates may be increased by some of these strategies, but can be filtered out with Picard (broadinstitute.github.io/picard).
If you see no peaks or enrichment in your Bioanalyzer/TapeStation traces, consider the following:
Are you analyzing CUT&RUN DNA yields are purified sequencing libraries?
- Remember, Bioanalyzer/TapeStation analysis is only to be conducted on prepared sequencing libraries and NOT raw CUT&RUN DNA.
- If you are analyzing sequencing libraries, read below.
Reasons for no peaks in traces derived from CUT&RUN libraries include:
- Low CUT&RUN yields and/or low inputs for library prep. See this article for guidance.
- Sub-optimal library prep technique. Use the EpiCypher CUTANA CUT&RUN Library Prep Kit (EpiCypher 14-1001 & 14-1002), specifically developed for CUT&RUN
No. While a few exceptions exist (e.g. RNA Pol II), the standard CUTANA™ CUT&RUN protocol is sufficient for the vast majority of protein targets tested by EpiCypher, including transcription factor motif analysis.
If you see large peaks (>500 bp) in your prepared libraries it is likely due to one of the following reasons:
Poor sample prep and/or incomplete permeabilization. Review quality control checks and reference sample prep steps and variations if applicable. Poor sample prep will impact the quality of your experimental sample. Additionally, if cells are not permeabilized properly, antibody and pAG-MNase cannot diffuse efficiently, leading to less target cutting and large fragment amplification. Optimize Digitonin permeabilization conditions for your cell type and repeat the CUT&RUN experiment. Alternatively, use nuclei for CUT&RUN, which do not require Digitonin optimization (use 0.01%).
Inefficient ConA bead binding. If cells are not bound to beads with >95% efficiency, you may observe higher background and larger fragment enrichment. If your cells are lysed after crosslinking, their binding efficiency will be low and leads to more clumping downstream as well. Check the percentage of cells/nuclei bound to the ConA beads. As above, working with nuclei may alleviate this issue.
PCR. Be sure to strictly follow the PCR steps as described in our protocol. The PCR parameters outlined are optimized for 200 – 700 bp fragments and should eliminate large fragments. If you are seeing enrichment of large fragments, be sure to check your PCR parameters.
These are adapter dimers, which result from self-ligation of sequencing adapters and preferential amplification during library prep (see Figure 1 below).
As long as you see predominant enrichment of mononucleosome-sized fragments at ~300 bp, and adapter dimers comprise less than 5% of your prepared library (i.e. % Integrated in trace is less than 5%), you can move forward with sequencing.
If you have >5% adapter dimers in your library, we recommend removing them via gel purification (see this FAQ).
If adapter dimers are the only peak you see we suggest repeating the experiment with more cells, and be sure to perform all quality control checks for sample prep. Include reactions with control antibodies and SNAP-CUTANA™ Spike-in Controls to validate your workflow.
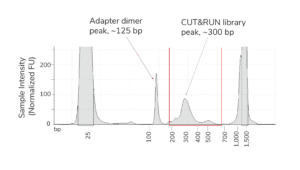
Figure 1. Example TapeStation trace from CUT&RUN H3K27me3 library containing an adapter dimer peak (~125 bp, red arrow) and expected library peak (~300 bp, blue arrow). Red lines denote the 200-700 bp range, used to determine library concentration.
Adapter dimers are generated by self-ligation of sequencing adapters that are preferentially amplified due to their small size. Adapter dimer contamination appears as a peak at ~125 bp (below), and is caused by low input DNA, inefficent adapter ligation, and/or using excess beads during library purification. Adapter dimers should represent no more than 5% of the total 200-700 bp fragment yield as determined on a Bioanalyzer/Tapestation trace (i.e. % Integrated in trace is less than 5%).
To minimize adapter dimers, keep adapter ligation reagents on ice during ligation setup. If adapter dimers are >5% of your library, they should be removed from libraries as outlined here.

Figure. Example TapeStation trace from CUT&RUN H3K27me3 library containing an adapter dimer peak (~125 bp, red arrow) and expected library peak (~300 bp, blue arrow). Red lines denote the 200-700 bp range, used to determine library concentration.
Adapter dimers should be removed when they comprise >5% of your library (i.e. % Integrated in trace is less than 5%), as determined by TapeStation or Bioanalyzer.
Adapter dimers can be removed by gel purification using the QIAquick Gel Extraction Kit (Qiagen 28704) or similar. It is recommended to gel purify the entire multiplexed library pool rather than gel-purifying individual libraries.
Gel purify DNA between 200-700 bp and cleanup as per manufacturer’s instructions. Reassess concentration and fragment distribution and proceed to sequencing. Additional reading about adapter dimers and library prep can be found in the CUTANA™ Library Prep Kit manual.
NO, you do not need to sequence an input control. Because there is no immunoprecipitation step in CUT&RUN, there is no input to sequence.
However, we do recommend including a reaction with the IgG negative control antibody, and sequencing this reaction in every experiment. This control will help gauge assay background.
Background in open chromatin may indicate digestion of accessible chromatin by pAG-MNase, due to prolonged incubation and/or incorrect temperature. However, it can also indicate over-sequencing of CUT&RUN libraries.
To resolve this:
- Sequence libraries to 3-8 million reads.
- Repeat assay using fresh buffers.
- Ensure MNase digestion is performed at 4ºC.
High background in sequencing data could indicate poor sample or library prep, over-digestion by MNase, DNA damage, or antibody failure.
To troubleshoot this, use 500,000 cells per reaction, including reactions with control antibodies and the K-MetStat Panel. Read the tips below:
- Confirm sample prep and include all quality control checks.
- Process cells quickly and resuspend in cold Antibody Buffer.
- Test multiple antibodies to experimental target.
- Ensure MNase digestion is incubated at 4ºC for 2 hours.
- Keep adapter ligation reagents on ice during ligation setup.
- Double check that library prep PCR parameters are accurate.
In general, ~1% of total sequencing reads should be assigned to spike-in controls. This sequencing bandwidth provides many thousands of spike-in reads, which is adequate to examine SNAP-CUTANA™ Spike-in recovery and/or to use E. coli reads for normalization. It also prevents spike-in reads from swamping sequencing data, ensuring that sufficient reads (3-8 million) are aligned to the species reference genome for biological analysis.
Note that the spike-in sequencing bandwidth may be higher or lower depending on target abundance, sequencing depth, and other factors. For instance, the IgG negative control often has 10-20% of reads assigned to the K-MetStat Panel, while a high abundance target (e.g. H3K27me3) may have 0.1-1%. Outside of this range, consider adjusting the spike-in dilution to be optimal for future experiments.