Thoroughly understanding disease mechanisms is key to successful drug discovery and development. Chromatin structure plays a significant role in disease and represents an important area of pharmaceutical research. As the genomics field grows, chromatin mapping assays – like EpiCypher’s CUTANA™ CUT&RUN Services – are increasingly valuable for pinpointing disease mechanisms and determining treatment impact. This blog explores the advantages of CUT&RUN for drug discovery and includes a case study illustrating the power of chromatin mapping assays to uncover druggable mechanisms.
Looking to boost your drug discovery and development pipelines? Here’s how
What is chromatin?
Chromatin is the biomolecular packaging of DNA in eukaryotic cells, which controls gene expression by modulating accessibility. Genes in “open” chromatin are more easily expressed, while “closed” chromatin is associated with gene silencing. Aberrant chromatin structure is linked to changes in gene expression across numerous diseases, including cancer, neurodegenerative diseases, developmental disorders, and autoimmunity.
Chromatin mapping assays are used to study the various proteins, enzymes, and chemical modifications that modulate chromatin accessibility and gene regulation. Histone post-translational modifications (PTMs) can mark genomic features like enhancers and promoters, offering valuable insights into gene expression mechanisms that are crucial for biomedical research.
How are chromatin mapping services used in drug discovery and development?
Chromatin mapping services and assays are critical tools to understand how gene expression is altered in disease. The results can help characterize new disease-relevant mechanisms, identify new drug targets, and monitor drug responses in (pre)clinical studies. In the era of personalized medicine, chromatin mapping may also be useful in matching patients to more effective drugs, resulting in more streamlined and effective treatment (Figure 1).
While traditional chromatin immunoprecipitation sequencing (ChIP-seq) is widely used, it has many pain points that make it unfit for pharmaceutical research, including poor sensitivity, low reliability, and slow turnaround.
Newer assays like Cleavage Under Targets and Release Using Nuclease (CUT&RUN) overcome these limitations. CUT&RUN is a rapid, ultra-sensitive chromatin mapping technique that generates more reliable data at higher resolution compared to ChIP-seq. With CUT&RUN, pharma companies can map chromatin interactions more precisely, accelerating drug discovery and improving clinical outcomes.
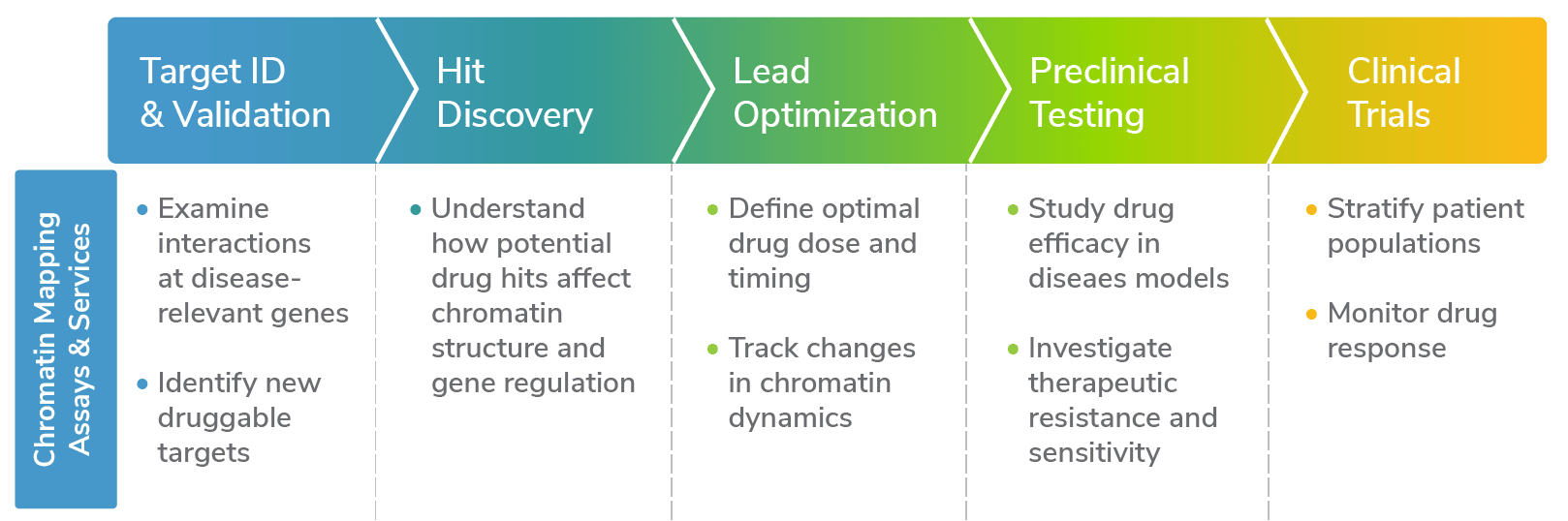
Figure 1: Chromatin mapping assays, such as CUTANA™ CUT&RUN kits and services, can be used throughout the drug development pipeline to support project success.
EpiCypher’s CUTANA CUT&RUN CUT&RUN Assays streamline this process further by providing high-throughput workflows compatible with primary cells and patient samples. Below, we highlight how CUTANA CUT&RUN contributed to identifying drug mechanisms in cancer research.
Case Study: Defining new drug mechanisms to overcome therapeutic resistance
Bagheri et al. Pharmacological induction of chromatin remodeling drives chemosensitization in triple-negative breast cancer. Cell Reports Medicine 2024.
The epithelial-to-mesenchymal transition (EMT) is a key driver of cancer growth, metastasis, and chemotherapy resistance, and is a defining feature of aggressive triple-negative breast cancer (TNBC) 1,2. In mouse models, disrupting or reversing EMT can sensitize breast cancer cells to chemotherapy, making it a key target in cancer drug research 3,4.
Unfortunately, translating these exciting results to targeted clinical therapies remains a challenge. Improved therapies require understanding how EMT and EMT reversal are modulated at the transcriptional level. The lack of sufficient chromatin mapping tools for large-scale projects and patient samples have greatly slowed these research efforts.
The approved chemotherapy drug eribulin has been found to modulate EMT in cancer cells 5. Here, Bagheri et al. investigated eribulin’s mechanism of action in TNBC cells lines, patient-derived xenograft models, and matched patient samples.
CUT&RUN Application
EpiCypher’s CUTANA CUT&RUN assays revealed that eribulin disrupts the interaction between EMT transcription factor ZEB1 and SWI/SNF chromatin remodelers, resulting in reduced ZEB1 binding at EMT genes. These chromatin changes directly correlated with improved chemotherapy response and reduced metastasis.
Advantages of CUT&RUN for this study
The high resolution and low background of CUTANA CUT&RUN made it possible to discern gradual changes in ZEB1 binding over a multi-day eribulin time course study. This is especially notable when compared to ChIP-seq assays, which often struggle to detect subtle changes in protein-protein interactions.
In addition, our streamlined CUT&RUN protocol only required 500,000 cells per reaction, making it possible to profile multiple targets from precious patient samples and xenograft models.
Key impact
This study highlights how chromatin mapping can uncover mechanisms of therapeutic resistance and inform drug development strategies for challenging cancers. Bagheri et al. found that eribulin – an FDA-approved drug – could be a useful tool in overcoming therapeutic resistance in aggressive TNBC. What other insights can epigenomics provide that will advance drug research?
What should you look for in a chromatin mapping services provider?
Drug developers may lack in-house expertise for chromatin mapping, making it essential to choose the right service provider. Here’s what to consider:
- Expertise: Look for a team with a strong track record in assay development and optimization.
- Support: Your provider should offer comprehensive support, from customized experimental design to preliminary data analysis.
- Scalability: Automation and high-throughput workflows are critical for scaling up large projects.
- High Sensitivity: CUT&RUN provides high-resolution data with minimal background, ensuring accurate and actionable results.
Can drug developers leverage EpiCypher CUT&RUN chromatin mapping services?
Yes! EpiCypher offers comprehensive CUT&RUN Services tailored to pharma needs. Our automated workflow is optimized for diverse sample types and scales easily, helping you generate high-quality data faster. With EpiCypher’s expertise, chromatin mapping can be useful at multiple points in drug development:
- Discovery of new druggable pathways regulating gene expression.
- Studying impact of drug and drug dosing in preclinical studies.
- Monitoring patient responses to tailor treatments.
- Identifying biomarkers to define drug efficacy.
Contact us today to discuss how we can support your drug development pipeline with CUT&RUN Services!
References
- Foulkes WD et al. Triple-negative breast cancer. N Engl J Med 363, 1938-48 (2010). https://doi.org/10.1056/NEJMra1001389.
- Nedeljković M et al. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells 8, (2019). https://doi.org/10.3390/cells8090957.
- Pattabiraman DR et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680 (2016). https://doi.org/10.1126/science.aad3680.
- Gupta PB et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645-59 (2009). https://doi.org/10.1016/j.cell.2009.06.034.
- Yoshida T et al. Eribulin mesilate suppresses experimental metastasis of breast cancer cells by reversing phenotype from epithelial-mesenchymal transition (EMT) to mesenchymal-epithelial transition (MET) states. Br J Cancer 110, 1497-505 (2014). https://doi.org/10.1038/bjc.2014.80.