Jackson ImmunoResearch now offers AffiniPure™ Rabbit Anti-GFP, an affinity-purified polyclonal antibody specific for GFP and its derivatives. The antibody’s polyclonal format enables spectacular signal amplification of expressed GFP. Learn how Jackson ImmunoResearch AffiniPure™ Rabbit Anti-GFP can be used to detect a GFP fusion protein expressed on yeast cells by flow cytometry.
Detection of GFP by flow cytometry using Anti-GFP antibodies
GFP is a 27kDa protein that emits green fluorescence when exposed to UV (ultraviolet) light. One of its derivatives, amongst others, is enhanced GFP (EGFP), which modifies the original jellyfish GFP (avGFP) to introduce two mutations (F64L, S65T) to improve brightness (Tsien, 1998).
EGFP is regularly used in fusion proteins as both an expression marker and as a reporter molecule. This is because it expresses as a functional, mature protein in most cell systems, doesn’t require additional cofactors, is easily visualized with common fluorescence microscopy set-ups, and is well validated making it reliable as a fusion protein. EGFP is regularly used as a reporter molecule to follow the gene expression in transiently transfected cells (Subramanian et al., 1996) and stable cell lines.
Although EGFP is a robust and tractable molecule, sample processing or experimental limitations can hinder its use in some conditions. We cover this below and introduce Anti-GFP antibodies as a solution to enable the use of GFP tags and bypass its limitations when conditions are suboptimal.
Limitations of GFP and its derivatives
Autofluorescence / background
Autofluorescence is intrinsic fluorescence that occurs due to the presence of cellular components. This fluorescence signal generates background that can interfere with the detection and interpretation of genuine signals from target proteins. Autofluorescence may occur across the spectrum but typically emits between 350 and 550 nm, which overlaps with the emission profile of GFP; this can be problematic as the GFP signal cannot be distinguished over the background autofluorescence. Anti-GFP antibodies conjugated with a fluorophore that emits in another, often longer, wavelength can circumvent these limitations by shifting GFP emission to another channel, and can then be distinguished from the autofluorescence.
Damage to the protein preventing luminescence
GFP, like many proteins, is sensitive to the conditions around them. For GFP to be able to fluoresce, it must form a fully folded beta-barrel structure. This is then followed by an intramolecular reaction that requires oxygen to generate the chromophore (Tsien, 1998). The consequence of this requirement is that GFP is non-fluorescent under anaerobic conditions. Another limitation of GFP that prevents its functionality is its sensitivity to acid. GFP’s chromophore can only absorb and emit light in its protonated state (Tsien, 1998). Therefore, at pHs below its pKa (<pH 6.0), where the chromophore is present in its deprotonated state, the efficiency of the protein to fluoresce is reduced.
Another issue with GFP, when expressed as a trans-protein under the promoter of a gene of interest, is that it may not be tethered within the cell and can diffuse within the cytosol. This may cause problems with subsequent immunostaining, where GFP can be lost during wash steps (Chalfie and Kain., 2005; Morris et al., 2010). Free movement of GFP can be prevented by fixing tissue/samples. However, this process, particularly FFPE, can functionally alter the GFP protein, weakening and destroying the protein’s ability to fluoresce (Kusser et al., 2003).
JIR Anti-GFP antibodies offer robust detection of GFP, recognizing GFP in either native or fixed/denatured conditions as long as the protein is not degraded, enabling the GFP tag to be used in circumstances where the protein would not be functional. The polyclonal format of JIR Rabbit Anti-GFP enables the binding of multiple antibodies to the target, bringing additional fluorescent molecules, thus enhancing the GFP signal beyond the output associated with GFP alone.
Flexible fluorescence
Another limitation of GFP relevant to flow cytometry is the choice of fluors. Often, flow cytometry uses fluorescent proteins such as Phycoerythrin, which provide robust and reliable signal. However, PE and GFP have overlapping spectral characteristics, making it difficult to excite the molecules separately and sufficiently deconvolute their signals from each other. Using an Anti-GFP antibody conjugated to a fluorophore with an emission profile in a longer wavelength, such as Alexa Fluor® 647, enables signals to be resolved without the need for spectral unmixing, which often requires additional or expensive equipment or additional data compensation. Conjugated Anti-GFP antibodies can also add versatility to experiments restricted by equipment limitations, for example, when a laser line or channel is occupied by the fluor of another target. Having the option to use different channels can be useful when labeling multiple targets.

Figure 1: PE and EGFP have similar spectral characteristics, which can cause overlap and prevent signal deconvolution.
Summary of the benefits of using an Anti-GFP antibody
- Retrieve/rescue and amplify GFP signal.
- Switch channel to circumvent background/ endogenous GFP signal and autofluorescence.
- Fluorescence flexibility – choose dyes to accommodate experimental needs
Case study: Expression confirmation: GFP tagged construct surface expression on yeast cells by flow cytometry.
Background:
Recombinant proteins can be expressed in various systems, including bacterial, insect, mammalian, and fungal/yeast. Recombinant proteins are regularly expressed as fusions with reporter molecules such as GFP to enable their detection, purification, or characterize interactions using such techniques as FRET (Förster resonance energy transfer). Flow cytometry is a useful technique for screening and quantifying expression, particularly when proteins are expressed on the surface of a cell. Here, we investigate the expression of EGFP fused to Aga2p, a yeast mating protein. Aga2p will form a heterodimer complex with Aga1p, resulting in EGFP display on yeast surface.

Figure 2: Schematic of Aga2p- EGFP fusion protein expressed on Yeast cells, detected by JIR Alexa Fluor® 647 Rabbit Anti-GFP.
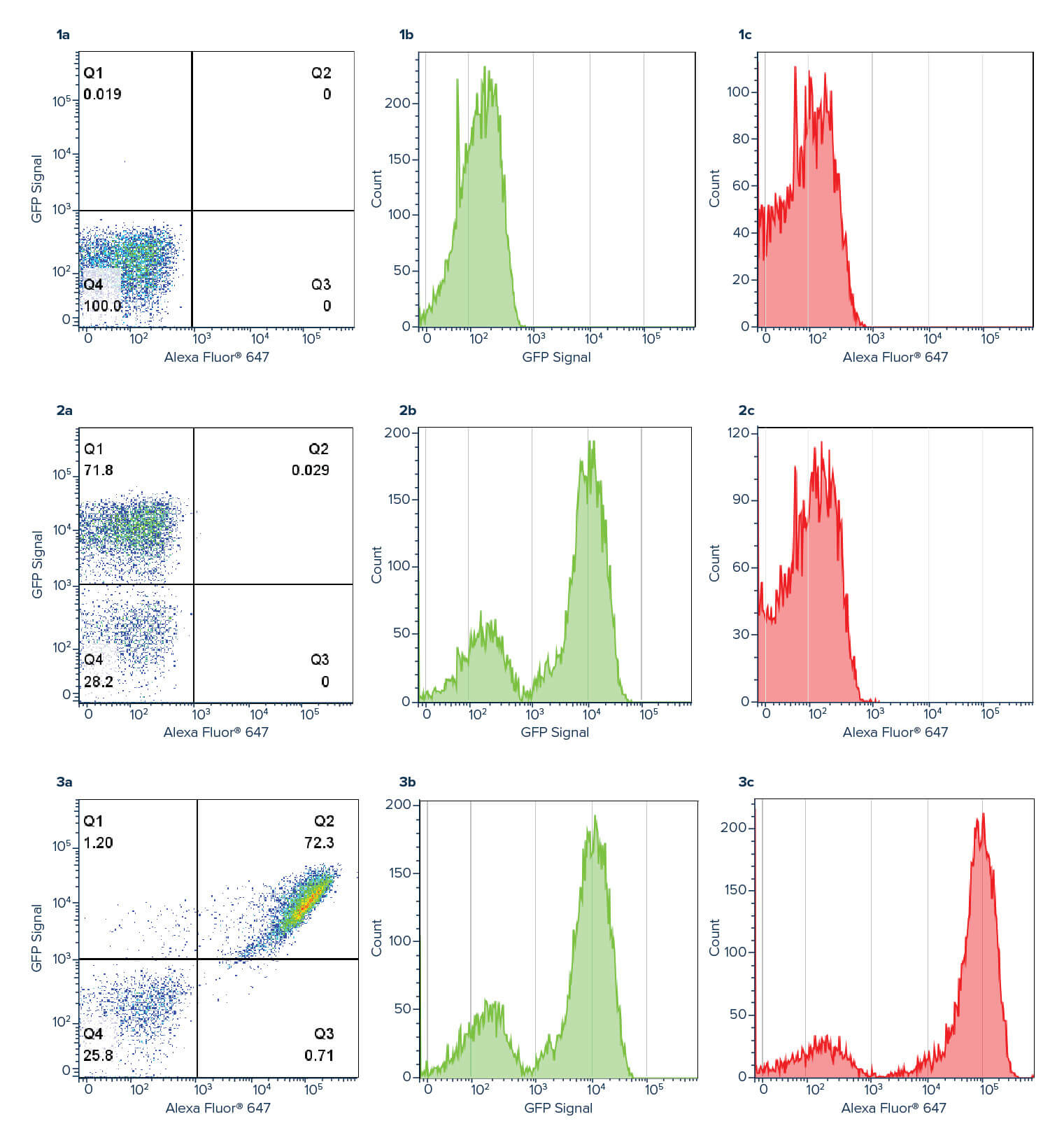
Figure 3:
Panel 1: Negative control. Yeast cells only, no GFP expression.
Panel 2: Yeast cells expressing EGFP fused to Aga2p. Panel 2b shows the GFP signal in the green channel. Panel 2c shows no signal in the red channel when no Alexa Fluor® 647 Anti-GFP is used.
Panel 3: Yeast cells expressing EGFP. Panel 3a Q2 shows double-labeled cells positive for signal in both green and red channels. Panel b shows the GFP signal in the green channel. Panel 3c shows Anti-GFP signal in the red channel. Amplification of the GFP signal can be seen when using Anti-GFP, with a log shift in brightness observed between the green and red channel.
Conclusion
Using an Anti-GFP antibody allows the GFP signal to be amplified, with a log shift in brightness observed between the green GFP channel (Figure 3b) and the red Alexa Fluor® 647 Anti-GFP channel (figure 3c). This amplification can help improve the deconvolution of data where channels may overlap, or the signal is indistinguishable from background.
Experimental set-up
EBY100 yeast (ATCC, MYA-4941) cells were transformed with a plasmid containing Aga2p-EGFP fusion gene using Frozen-EZ yeast transformation II kit (Zymo Research). Yeast cells were grown in SDCAA media (Teknova) lacking tryptophan as a selection marker at 30°C and 200 r.p.m. Protein expression was induced with 1:10 dilution of yeast grown in SDCAA into SGCAA media (Teknova) overnight at 30°C. The next day, 200 uL cells were washed with PBS + 0.2% BSA and pelleted before staining with Alexa Fluor® 647 Rabbit Anti-GFP for one h at room temperature (RT) followed by a wash step before resuspension in PBS + 0.2% BSA. Flow cytometry was performed using BD FACSCelesta and BD FACSDiva software (BD Biosciences). Analysis was performed using FlowJo software. Method adapted from Butler et al., 2022.
References
- Butler, Y.R., Liu, Y., Kumbhar, R., et al. α-Synuclein fibril-specific nanobody reduces prion-like α-synuclein spreading in mice. Nat Commun 13, 4060 (2022). https://doi.org/10.1038/s41467-022-31787-2
- Chalfie, M. & Kain, S. R. Green Fluorescent Protein: Properties, Applications, and Protocols: Second Edition. vol. 47 (Wiley Blackwell, 2005)
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., & Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science (New York, N.Y.), 263(5148), 802-805. https://doi.org/10.1126/science.8303295
- Chalfie, M. (1995). GREEN FLUORESCENT PROTEIN. Photochemistry and Photobiology, 62: 651-656. https://doi.org/10.1111/j.1751-1097.1995.tb08712.x
- Falkow, S., Valdivia, R., Cormack, B.,(1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene,173, 1, 33-38, https://doi.org/10.1016/0378-1119(95)00685-0
- Kusser, K. L., & Randall, T. D. (2003). Simultaneous detection of EGFP and cell surface markers by fluorescence microscopy in lymphoid tissues. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society, 51(1), 5-14. https://doi.org/10.1177/002215540305100102
- Lippincott-Schwartz, J., Snapp, E., & Kenworthy, A. (2001). Studying protein dynamics in living cells. Nature reviews. Molecular cell biology, 2(6), 444-456. https://doi.org/10.1038/35073068
- Morris, L. M., Klanke, C. A., Lang, S. A., Lim, F. Y., & Crombleholme, T. M. (2010). TdTomato and EGFP identification in histological sections: insight and alternatives. Biotechnic & histochemistry : official publication of the Biological Stain Commission, 85(6), 379-387. https://doi.org/10.3109/10520290903504753
- Scandella, V., Paolicelli, R. C., & Knobloch, M. (2020). A novel protocol to detect green fluorescent protein in unfixed, snap-frozen tissue. Scientific reports, 10(1), 14642. https://doi.org/10.1038/s41598-020-71493-x
- SHIMOMURA, O., JOHNSON, F. H., & SAIGA, Y. (1962). Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. Journal of cellular and comparative physiology, 59, 223-239. https://doi.org/10.1002/jcp.1030590302
- Snapp E. (2005). Design and use of fluorescent fusion proteins in cell biology. Current protocols in cell biology, Chapter 21, 21.4.1-21.4.13. https://doi.org/10.1002/0471143030.cb2104s27
- Soboleski, M. R., Oaks, J., & Halford, W. P. (2005). Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 19(3), 440-442. https://doi.org/10.1096/fj.04-3180fje
- Stretton, S., Techkarnjanaruk, S., McLennan, A. M., & Goodman, A. E. (1998). Use of green fluorescent protein to tag and investigate gene expression in marine bacteria. Applied and environmental microbiology, 64(7), 2554-2559. https://doi.org/10.1128/AEM.64.7.2554-2559.1998
- Subramanian, S., & Srienc, F. (1996). Quantitative analysis of transient gene expression in mammalian cells using the green fluorescent protein. Journal of biotechnology, 49(1-3), 137-151. https://doi.org/10.1016/0168-1656(96)01536-2
- Tsien R. Y. (1998). The green fluorescent protein. Annual review of biochemistry, 67, 509-544. https://doi.org/10.1146/annurev.biochem.67.1.509
