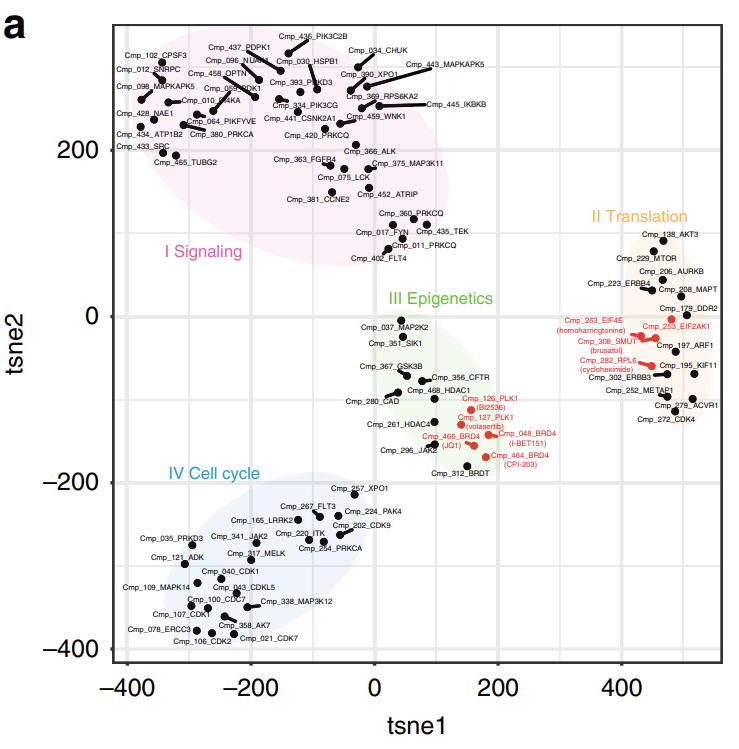
Figure 1. t-distributed Stochastic Neighbor Embedding (t-SNE) clustering using 4289 dysregulated genes following compound treatment at 10 μM. Colors group compounds with common mechanisms of action. Each compound and its target are labeled. Figure is taken from Ye et al. (2018) Nature Communications.
In the translation cluster, two compounds that targeted key components of the translation machinery clustered closely with a compound with unknown targets. This suggested that the compound with unknown targets likely shared a similar mechanism of action.
In the epigenetic cluster, related but distinct compounds were grouped together. Unexpectedly, these compounds had different dose-dependent effects on gene expression.
Together, these results highlight that DRUG-seq can be used to infer mechanisms of action, potential targets of novel compounds, and their dose dependencies.
As further validation of their technique, Ye et al. (2018) compared their results to the Connectivity Map database; a fixed panel of about 1000 landmark genes following compound treatment (Subramanian et al., 2017). They confirmed that 52 of the 433 compounds causing gene expression changes were also present in this independent data set.
Validation for CRISPR knockout cells
Many drug discovery applications now begin with CRISPR/CAS9 gene editing, especially when target-specific compounds are unavailable. Therefore, Ye et al. (2018) compared the transcriptional effects of CRISPR knockout and compound treatment. They did this for a key component of the translation machinery called RPL6.
DRUG-seq found that CRISPR reduced RPL6 mRNA levels but drug treatment did not. Furthermore, CRISPR- and compound-treated cells had only a partial overlap in differentially expressed genes.
This was possibly due to the different mechanisms used by CRISPR or compound treatment to inhibit RPL6. The different genes detected also indicated possible off-target effects for each method. Therefore, DRUG-seq is a powerful tool for whole transcriptome readouts in a high-throughput compound or CRISPR screening environment.
Other high-throughput transcriptomic methods for drug discovery
Other technologies such as Bulk RNA Barcoding and sequencing (BRB-seq) are also extremely well suited to high-throughput transcriptomic screening of CRISPR- or compound libraries in the industrial setting. Of note, BRB-seq has been extensively optimized and validated to provide sensitive, accurate data even for low-quality RNA (Alpern et al., 2018).
Furthermore, Alithea Genomics has extensively validated BRB-seq to give the best quality data for different cell types, such as blood cells, with the inclusion of a proprietary globin blocker.
Thanks to these technologies, researchers and the pharmaceutical industry can now screen and infer the likely targets, potential mechanisms of action, and subtle dose dependencies for thousands of compounds with unparalleled accuracy.
Please contact us at info@stratech.co.uk to find out more about how BRB-seq could help in your high-throughput drug discovery study.
References:
- Alpern, D., Gardeux, V., Russeil, J., Mangeat, B., Meireles-Filho, A.C., Breysse, R., Hacker, D. and Deplancke, B., 2019. BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome biology, 20(1), pp.1-15.
- Li, J., Ho, D.J., Henault, M., Yang, C., Neri, M., Ge, R., Renner, S., Mansur, L., Lindeman, A., Kelly, B. and Tumkaya, T., 2022. DRUG-seq Provides Unbiased Biological Activity Readouts for Neuroscience Drug Discovery. ACS Chemical Biology. 17(6), pp.1401-1414.
- Subramanian, A., Narayan, R., Corsello, S.M., Peck, D.D., Natoli, T.E., Lu, X., Gould, J., Davis, J.F., Tubelli, A.A., Asiedu, J.K. and Lahr, D.L., 2017. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell, 171(6), pp.1437-1452.
- Ye, C., Ho, D.J., Neri, M., Yang, C., Kulkarni, T., Randhawa, R., Henault, M., Mostacci, N., Farmer, P., Renner, S. and Ihry, R., 2018. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nature communications, 9(1), pp.1-9.