An ongoing outbreak of a novel coronavirus (SARS-CoV-2) has raised global concerns. It is identified as the cause of pneumonia with unknown etiology. Since the early outbreak in Wuhan, China, it has subsequently spread to all provinces of China and many other countries. The urgent epidemic situation has spurred the development of antiviral drugs and vaccines. As a leading service provider in the field of biological research and drug discovery, Creative Biolabs provides fast & elaborate therapeutic antibody discovery, drug candidates screening and vaccine development services to help combat this outbreak.
Classification of SARS-CoV-2
Using unbiased sequencing and isolation from patients’ samples, SARS-CoV-2 was identified as another clade within the Betacoronavirus genus, Coronaviridae family. The two other strains of Betacoronaviruses-severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV)—have caused more than 10,000 cumulative cases in the past two decades, with mortality rates of 10% for SARS-CoV and 37% for MERS-CoV. Similar to SARS-CoV and MERS-CoV infections, patients with SARS-CoV-2 also exhibited symptoms of viral pneumonia including fever, dyspnea, bilateral lung infiltration, respiratory failure, even death.
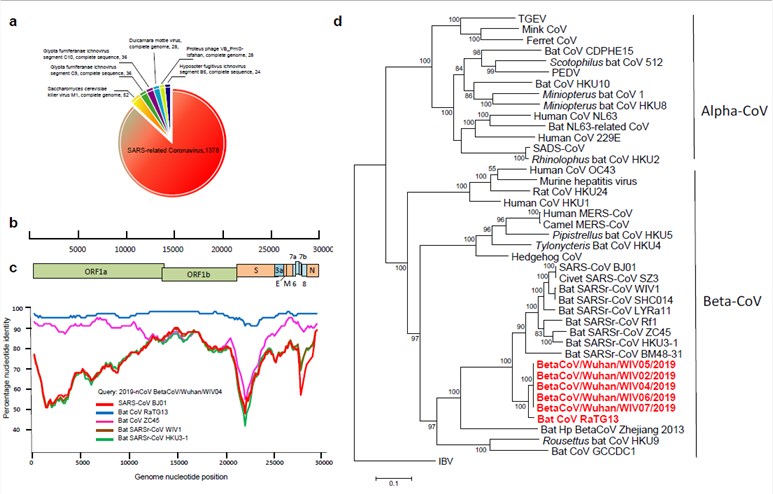
Genome Characterization of SARS-CoV-2
With the help of high throughput sequencing and metagenomic analysis, scientists have obtained full-length genome sequences from five patients at the early stage of the outbreak. To date, a series of SARS-CoV-2 sequences have been uploaded and are available in GenBank and the Sequence Read Archive (SRA).
Downloaded website: https://www.ncbi.nlm.nih.gov/genbank/SARS-CoV-2-seqs/
All of the sequences are almost identical to each other and share 79.5% sequence identity to SARS-CoV. Furthermore, SARS-CoV-2 shows 96% identity at the whole genome level to a bat coronavirus. The pairwise protein sequence analysis of seven conserved non-structural proteins shows that this virus belongs to the species of SARSr-CoV.
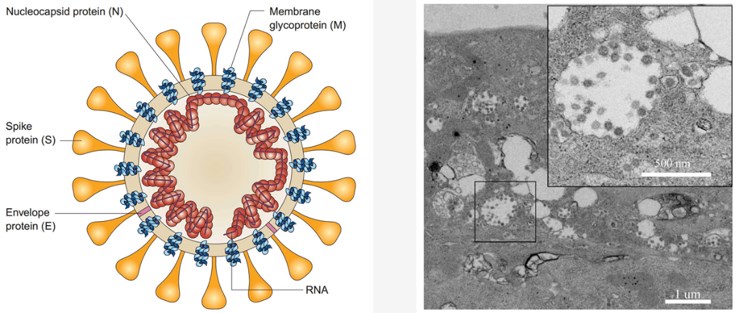
Structure of Coronavirus
Coronaviruses are enveloped viruses with positive sense, single-stranded RNA genomes. Its genome size ranges from 26 to 32 kilobases (kb) in length and encodes a large polyprotein (ORF1a/b) that is proteolytically cleaved to generate 15 or 16 non-structural proteins. The 3’ end of coronaviral genome encodes four major structural proteins: the spike (S) protein, nucleocapsid (N) protein, membrane (M) protein, and the envelope (E) protein. According to detailed genomic and structure-based analysis, SARS-CoV-2 shares several functionally important components: ORF1a, ORF1b, and major structural proteins S, N, M E.
- S protein normally mediates virus entry into host cells. It facilitates the attachment to the host cell surface receptors and subsequent fusion between the viral and host cell membranes. In addition to mediating virus entry, the S protein is a critical determinant of viral host range and tissue tropism and a major inducer of host immune responses.
- N protein functions primarily to bind to the RNA genome, making up the nucleocapsid.
- M protein defines the shape of the viral envelope. It is also regarded as the central organizer of virus assembly, interacting with E protein for virus production and release.
- E protein mainly participates in virus assembly, maturation, and budding.
Scientists have confirmed that SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as the cell entry receptor, similar to SARS-CoV.
Diagnostic Methods of SARS-CoV-2
- Clinical symptoms should be used to guide testing: fever, cough, and shortness of breath.
- Imaging diagnosis (CT): a key component in diagnosing SARS-CoV-2, which usually shows ground-glass lesions with “white lung-like” changes.
- RT-PCR: primary and secondary RT-PCR assays are still golden standards for the diagnosis and confirmation of SARS-CoV-2.
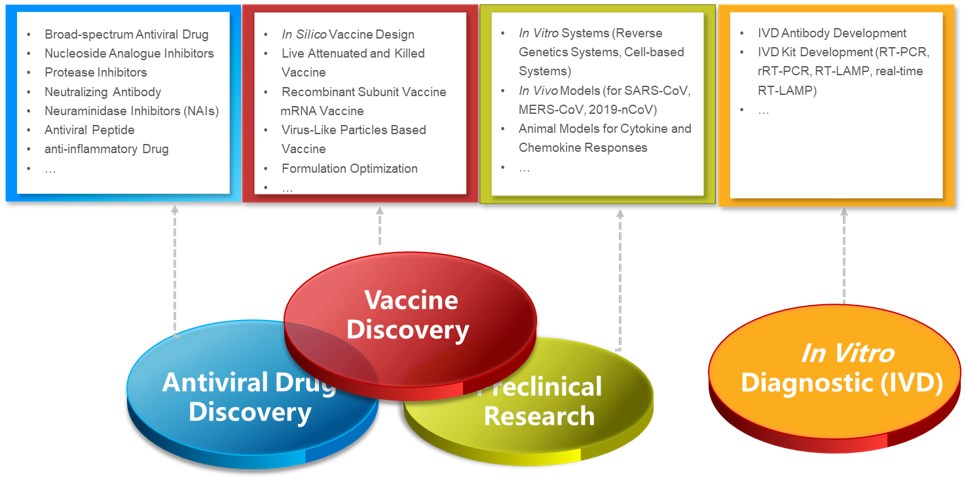
Services at Creative Biolabs
The severity and emergency of SARS-CoV-2 infection spurs an urgent need for antiviral drug and vaccine discovery. Creative Biolabs has spent considerable efforts to provide scientific support and comprehensive services to promote the success of this combat. Equipped with cutting-edge techniques and vast experience, we are keeping up with current research hotspots and integrating all resources to guarantee high-quality services (including but not limiting to): Antiviral Drug Discovery, Vaccine Discovery, Preclinical Research, In Vitro Diagnostic (IVD) Immunoassays.
References
- Zhou, P.; et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020.
- Gralinski, L. E.; Menachery, V. D. Return of the Coronavirus: 2019-nCoV. Viruses. 2020, 12(2): 135.
- Li, Q.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. New England Journal of Medicine. 2020.
 For Research Use Only. We do not provide services or products directly for patients.
For Research Use Only. We do not provide services or products directly for patients.