Creative Diagnostics now offers immunoassay kits for direct detection of the new coronavirus and antibodies against SARS-CoV-2
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, previously named 2019 novel coronavirus or 2019-nCoV) disease (COVID-19) from China at the end of 2019 has become a global pandemic and an unprecedented public health issue.
Creative Diagnostics started developing a new test shortly after the outbreak had become known. Only a few weeks later, two ELISAs for the detection of Anti-SARS-CoV-2 antibodies of classes IgM and IgG and one ELISA for the detection of SARS-CoV-2 NP antigen were available. Both test systems are currently only available to research institutions as research-use-only products. Preparations for CE registration of these tests are underway.
CREATIVE DIAGNOSTICS COVID-19
Research Solutions for Coronavirus – Creative Diagnostics
SARS-CoV-2 Immunoassay Kits
Technical Specifications
DEIASL019 SARS-CoV-2 IgG ELISA Kit
| Principle | Indirect ELISA |
| Sample Type | Serum/Plasma |
| Sample Volume | 10 uL |
| Incubation Time | 50 mins, 37 °C |
| Total Wash Steps | 2 |
Principles of Testing: This kit is based on the principle of indirect method (ELISA) to detect the SARS-COV-2 IgG antibody in human serum or plasma. The SARS-COV-2 whole virus lysate antigen is pre-coated on the enzyme-labeled strips. Add the test sample and incubate it. The IgG antibody in the sample is bound to the antigen. Wash the plate to remove SARS-COV-2 IgG that does not bind to the coated antigen. Add the enzyme-labeled reagent for a second incubation. When a SARS-COV-2 IgG antibody is present in the sample, a “coated antigen-IgG antibody-anti-human IgG enzyme conjugate” complex will be formed. After washing the plate again, color reagent is added, and the HRP linked to the complex will catalyze the color The reagent reacts to produce a blue product, which turns yellow after the reaction is terminated; if no SARS-COV-2 IgG antibody is present in the sample, it does not develop a color. Measure the OD value on a microplate reader or enzyme immunoassay system, and determine the presence or absence of a SARS-COV-2 IgG antibody based on the OD value
DEIASL020 SARS-CoV-2 IgM ELISA Kit
| Principle | Capture ELISA |
| Sample Type | Serum/Plasma |
| Sample Volume | 10 uL |
| Incubation Time | 80 mins, 37 °C |
| Total Wash Steps | 2 |
Principles of Testing: This kit is based on the capture ELISA to detect the SARS-COV-2 IgM antibody in human serum or plasma. The anti-µ chain monoclonal antibody is pre-coated on the microplate wells. Add the test sample and incubate it. The IgM antibody in the sample is bound to anti-µ chain monoclonal antibody. Wash the plate to remove SARS-COV-2 IgM that does not bind to the microplate wells. Add the enzyme-labeled reagent for a second incubation. When a SARS-COV-2 IgM antibody is present in the sample, a “anti-µ chain monoclonal antibody-human anti SARS-CoV-2 IgM antibody-SARS-CoV-2 antigen enzyme conjugate” complex will be formed.
DEIA2020 SARS-CoV-2 Antigen ELISA Kit
| Principle | Sandwich ELISA |
| Sample Type | Serum |
| Sample Volume | 25 uL |
| Incubation Time | 90 mins, 37 °C |
| Total Wash Steps | 2 |
Principles of Testing: The new Coronavirus (SARS-COV-2) antigen detection kit is based on the principle of a double antibody sandwich enzyme-linked immunoassay. The microplate is pre-coated with an anti-new coronavirus N protein antibody. When the serum and plasma to be detected contain the new coronavirus N protein antigen, it will bind to the specific antibody coated on the microplate. After the HRP labeled detection antibody against the new coronavirus N protein was added, an “antibody-antigen-enzyme labeled antibody” complex is formed. After adding the chromogenic substrate TMB, HRP enzyme will catalyze the color development. After terminating the solution, the microplate reader will determine the Absorbance value to determine the presence or absence of new coronavirus antigen in each test sample.
SARS-CoV-2 Antigens and Antibodies
SARS-CoV-2 (2019-nCoV) is a new type of bat coronavirus identified as the cause of an outbreak of respiratory illness first detected in Wuhan, China. Early on, many of the patients in the outbreak in Wuhan, China reportedly had some link to a large seafood and animal market, suggesting animal-to-person spread. However, a growing number of patients reportedly have not had exposure to animal markets, indicating person-to-person spread is occurring. At this time, it’s unclear how easily or sustainably this virus is spreading between people.
The coronavirinae family consists of four genera based on their genetic properties, including genus Alphacoronavirus, genus Betacoronavirus, genus Gammacoronavirus, and genus Deltacoronavirus. Host range and tissue tropism show a lot of variation among different CoVs. Generally, the Alphacoronavirus and Betacoronavirus can infect mammals, and the Gammacoronavirus and Deltacoronavirus can infect birds, but some of them can also infect mammals. SARS-CoV-2 (2019-nCoV), SARS coronavirus and MERS coronavirus all belong to the Betacoronavirus genus and are zoonotic pathogens that can cause severe respiratory diseases in humans.
Genome analysis of SARS-CoV-2 (2019-nCoV) showed that SARS-CoV-2 (2019-nCoV) was slightly different from SARS CoV and MERS CoV, but the functionally important ORFs, ORF1a and ORF1b, and major structural proteins such as the spike (S), membrane (M), envelop (E) and nucleic capsid (N) proteins are well annotated. Four structural proteins are essential for virion assembly and infection of CoVs. Homotrimers of S proteins make up the spike on the surface of virus particles and it is the sole viral membrane protein responsible for cell entry. It binds to the receptor on the target cell and mediates subsequent virus-cell fusion. It is also the key target for vaccine design. The M protein has three transmembrane domains and shapes the virions, promotes membrane curvature, and binds to the nucleocapsid. The E protein plays a role invirus assembly and release, and it is required for pathogenesis. The N protein contains two domains, both of them can bind virus RNA genome via different mechanisms. It is reported that N protein is an antagonist of interferon and viral encoded repressor (VSR) of RNA interference (RNAi), which benefit the viral replication.
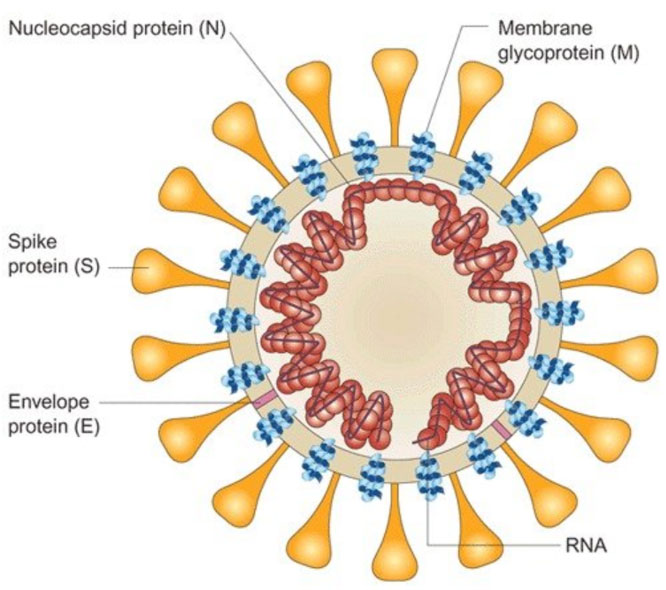
SARS-CoV-2 (2019-nCoV) spike protein, which belongs to class I virus fusion protein, contains two subunits, S1 and S2. S1 mainly contains receptor binding domain (RBD), which is responsible for identifying cell receptors. S2 contains the basic elements needed for the membrane fusion process. It has been reported that SARS-CoV-2 (2019-nCoV) spike protein interacts with human ACE2 to infect human respiratory epithelial cells.
Creative Diagnostics now can provide recombinant SARS-CoV-2 (2019-nCoV) spike, nucleocapsid antigens. These antigens can be used for scientific research, detection of novel coronavirus and preparation of neutralizing antibodies. All of our antigens are produced using a standardized production process to ensure the highest quality and are performance guaranteed for the applications listed on the detailed datasheets.
SARS-CoV-2 Antigens
| Target | Cat.No. | Product Name | Host |
| S Protein | DAGC158 | Recombinant SARS-CoV-2 Spike(S1+S2 ECD) Protein [His] | Insect Cells |
| DAGC159 | Recombinant SARS-CoV-2 Spike Protein [His] | Yeast | |
| S1 Protein | DAGC091 | Recombinant SARS-CoV-2 Spike Protein 1 [His] | HEK293 |
| DAGC092 | Recombinant SARS-CoV-2 Spike Protein 1 [human Fc] | HEK293 | |
| DAGC093 | Recombinant SARS-CoV-2 Spike Protein 1 [mouse Fc] | HEK293 | |
| DAGC138 | Recombinant SARS-CoV-2 Spike Protein 1 | HEK293 | |
| DAGC157* | Recombinant SARS-CoV-2 Spike 1 Protein (truncated, 30 KDa) [His] | E. coli | |
| S RBD Protein | DAGC089 | Recombinant SARS-CoV-2 Spike Protein Receptor Binding Domain [Fc] | HEK293 |
| DAGC149 | Recombinant SARS-CoV-2 Spike Protein Receptor Binding Domain [His] | HEK293 | |
| S2 Protein | DAGC153 | Recombinant SARS-CoV-2 S2 Protein (S2 ECD) [His] | Insect Cells |
| Nucleocapsid Protein | DAGC094 | Recombinant SARS-CoV-2 Nucleocapsid Protein (full length) [His] | E. coli |
| DAGS029 | Recombinant SARS-CoV-2 Nucleocapsid Protein (29-213 aa) [His] | E. coli | |
| DAGS059 | Recombinant SARS-CoV-2 Nucleocapsid Protein (59-213 aa) [His] | E. coli | |
| DAGC140 | Recombinant SARS-CoV-2 Nucleocapsid protein N3 fragment (C-ter domain, 28 KDa) | E. coli | |
| DAGC141 | Recombinant SARS-CoV-2 Nucleocapsid protein N5 fragment (N-ter domain, 43 KDa) | E. coli | |
| DAGC155* | Recombinant SARS-CoV-2 Nucleocapsid Protein (truncated, N3) [His] | E. coli | |
| DAGC156* | Recombinant SARS-CoV-2 Nucleocapsid Protein (truncated, 28 KDa) [His] | E. coli | |
| NSP3 Protein | DAGC154 | Recombinant SARS-CoV-2 papain-like protease [His] | E. coli |
*Highly recommended for the SARS-CoV-2 antibody lateral flow immunoassay development.
SARS-CoV-2 Antibodies
| Target | Cat. No. | Product Name | Application | Isotype | Capture-Detection |
| S Protein | CABT-CS031 | Human Anti-SARS-CoV-2 S1 Monoclonal antibody, clone BIB112 | ELISA | IgG | CABT-CS031 – CABT-CS033 |
| CABT-CS032 | Human Anti-SARS-CoV-2 Spike RBD Monoclonal antibody, clone BIB113 | ELISA | IgG | ||
| CABT-CS033 | Human Anti-SARS-CoV-2 S1 Monoclonal antibody, clone BIB114 | ELISA | IgG | CABT-CS031 – CABT-CS033 | |
| CABT-CS034 | Human Anti-SARS-CoV-2 Spike RBD Monoclonal antibody, clone BIB115 | ELISA | IgG | ||
| CABT-CS035 | Mouse Anti-SARS-CoV-2 Spike RBD Monoclonal antibody, clone 211184 | ELISA | IgG | ||
| CABT-CS044 | Human Anti-SARS-CoV-2 Spike RBD Monoclonal antibody, clone BIB116 | ELISA | IgM | ||
| CABT-RM321 | Rabbit anti-SARS-CoV-2 spike glycoprotein monoclonal antibody, clone 118 | ELISA | IgG | ||
| NP | CABT-CS026 | Mouse Anti-SARS-CoV-2 NP Monoclonal antibody, clone 7G21 | ELISA, WB | IgG | |
| CABT-CS027 | Mouse Anti-SARS-CoV-2 NP Monoclonal antibody, clone 4B21 | ELISA, IHC | IgG | ||
| CABT-CS024 | Rabbit Anti-SARS-CoV-2 NP Polyclonal antibody | ELISA | IgG | ||
| CABT-RM320 | Rabbit anti-SARS-CoV-2 NP monoclonal antibody, clone 120 | ELISA, WB | IgG | CABT-RM320 – CABT-CS037 | |
| CABT-CS037 | Human Anti-SARS-CoV-2 Nucleoprotein Monoclonal antibody, clone CO10 | ELISA | IgG | CABT-RM320 – CABT-CS037 |
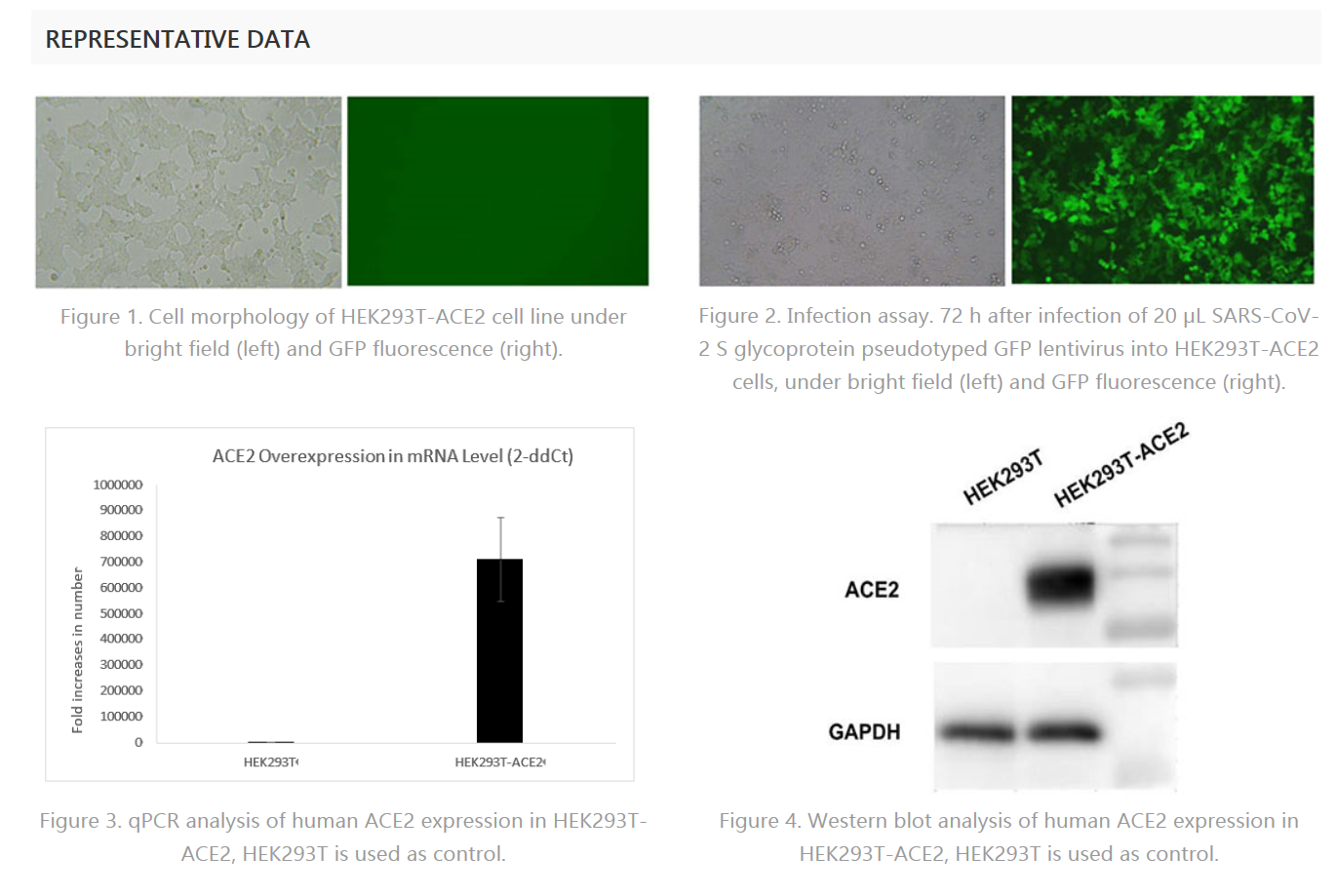
Human ACE2 Expressing Stable Cell Line
| Supplier | Product name | code | Expression Host |
| Creative Diagnostics | Human ACE2 Expressing Stable Cell Line | CSC-ACE01 | HEK293T |
This cell line is constructed by transduction of human angiotensin I converting enzyme 2 (ACE2) into HEK293T cells, followed by stable cell selection. HEK293T is derived from HEK293 and is commonly used in scientific research. HEK293T-human ACE2 cell line can be used for in vitro screening and characterization of drug candidates against SARS-CoV, SARS-CoV-2 (2019-nCoV).