Reliable and robust chromatin mapping assays
- Extremely reliable, cost-effective workflow
- Fewer cells required
- Only 3-5 million reads required / sample
- High signal : noise ratio
- Diverse sample inputs and targets
CUTANA™ CUT&RUN offers distinct advantages vs. ChIP-seq, the standard chromatin mapping assay.
The Cleavage Under Targets and Release Using Nuclease (CUT&RUN) method builds upon Chromatin ImmunoCleavage (ChIC) technology.
In CUT&RUN, a fusion of protein A, protein G and micrococcal nuclease (pAG-Mnase) is used to selectively cleave antibody-labelled chromatin. This strategy eliminates immunoprecipitation steps, greatly simplifying the assay workflow. Clipped chromatin fragments are isolated from solution and used for NGS.
The targeted enrichment of CUT&RUN allows for better signal : noise with only 3-5 million reads, significantly reducing sequencing costs vs ChIP-seq.
CUT&RUN has superior signal : noise with
> 10-fold reduced seq depth compared to ChIP-seq
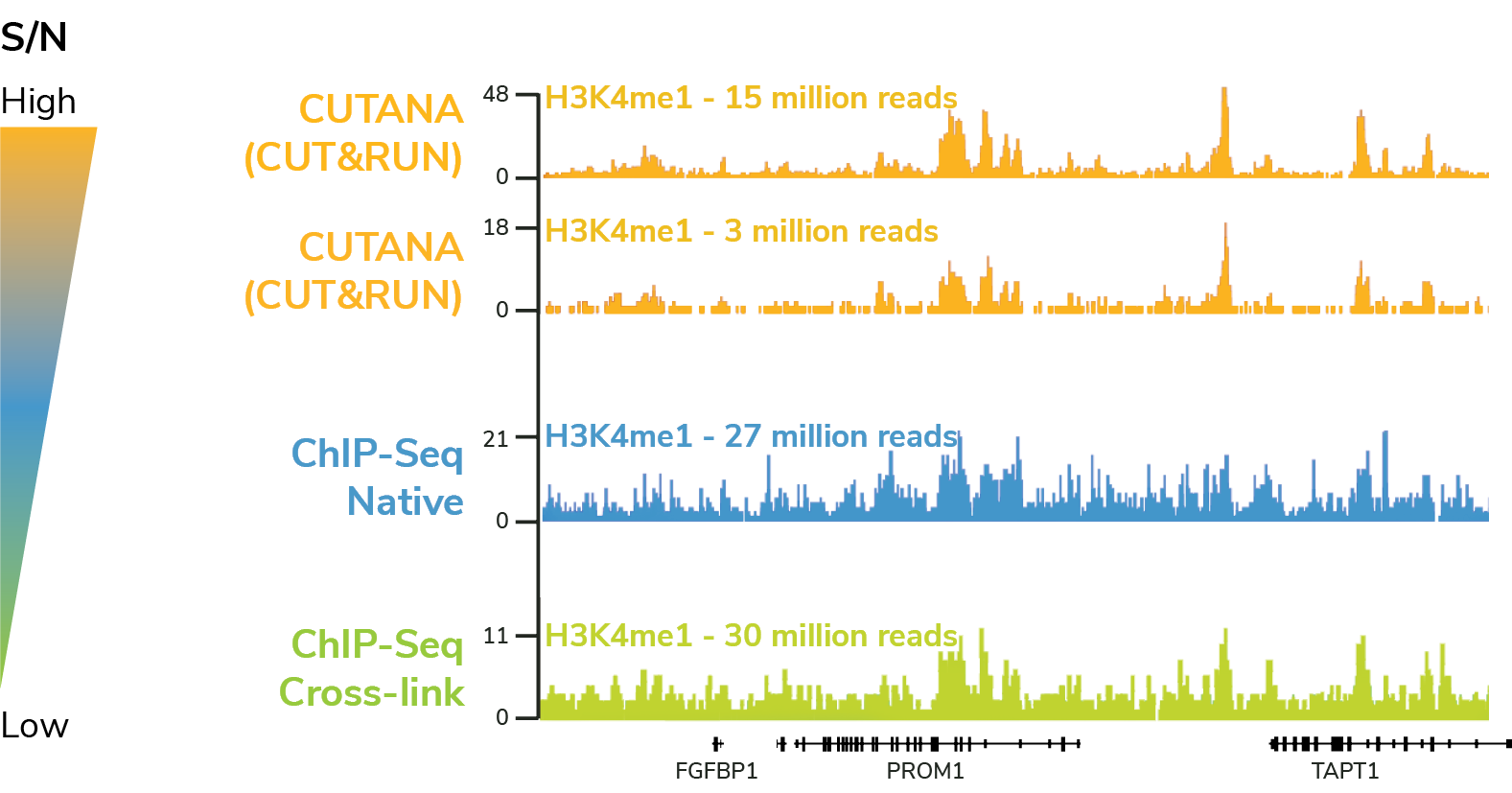
A representative 350 kb region of H3K4me1 profiles in K562 cells, generated using CUT&RUN (yellow tracks), native ChIP-seq (blue tracks), or cross-linked ChIP-seq (green tracks). All data were generated by EpiCypher and are expressed as reads per million (RPM). Color-coded gradient (to right) represents signal-to-noise ratios determined by genome-wide analysis (bamFingerprint data, not shown).
CUTANA™ CUT&RUN:
Go from cells to data in < 4 Days
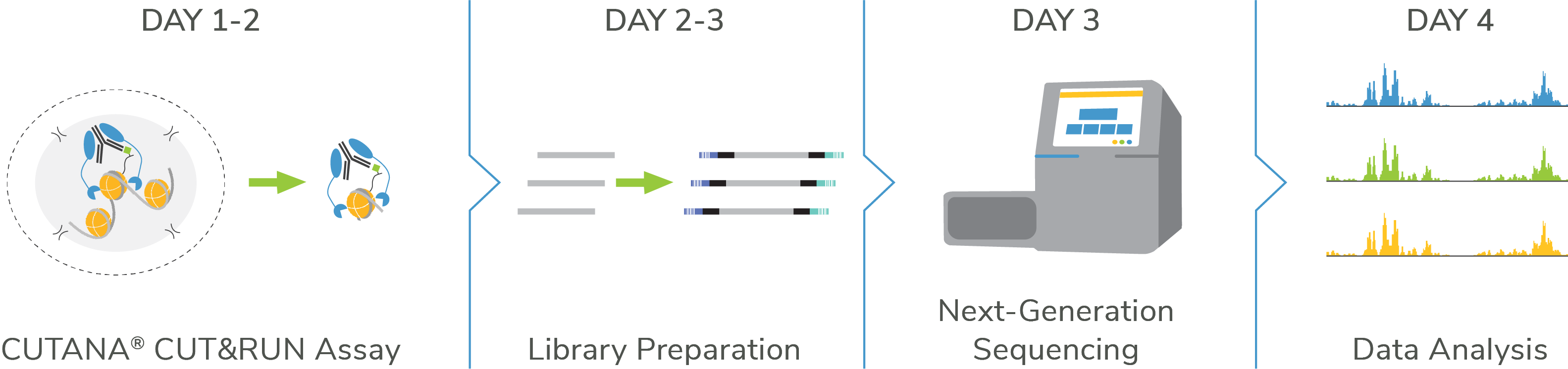
Get started with CUTANA™ CUT&RUN assays

Start performing your own CUT&RUN experiments with CUTANA pAG-MNase, the key reagent for CUT&RUN assays. Available for 50 or 250 reactions.
SNAP-ChIP™ certified, CUTANA-compatible antibodies are rigorously validated to ensure comparable results in ChIP-seq and CUT&RUN assays.
EpiCypher offers a collection of products for CUT&RUN assays, including pAG-MNase, spike-in controls, antibodies, and accessory reagents / tools, making it easy to create your own assay or follow a standard protocol.
We have developed a reliable and user-friendly CUT&RUN protocol, optimized for:
In addition, we have addressed common issues (e.g. bead clumping / loss) and enabled high-throughput sample handling via an 8-strip format. The protocol includes notes throughout, critical steps, pictures for clarity, and a FAQ section.