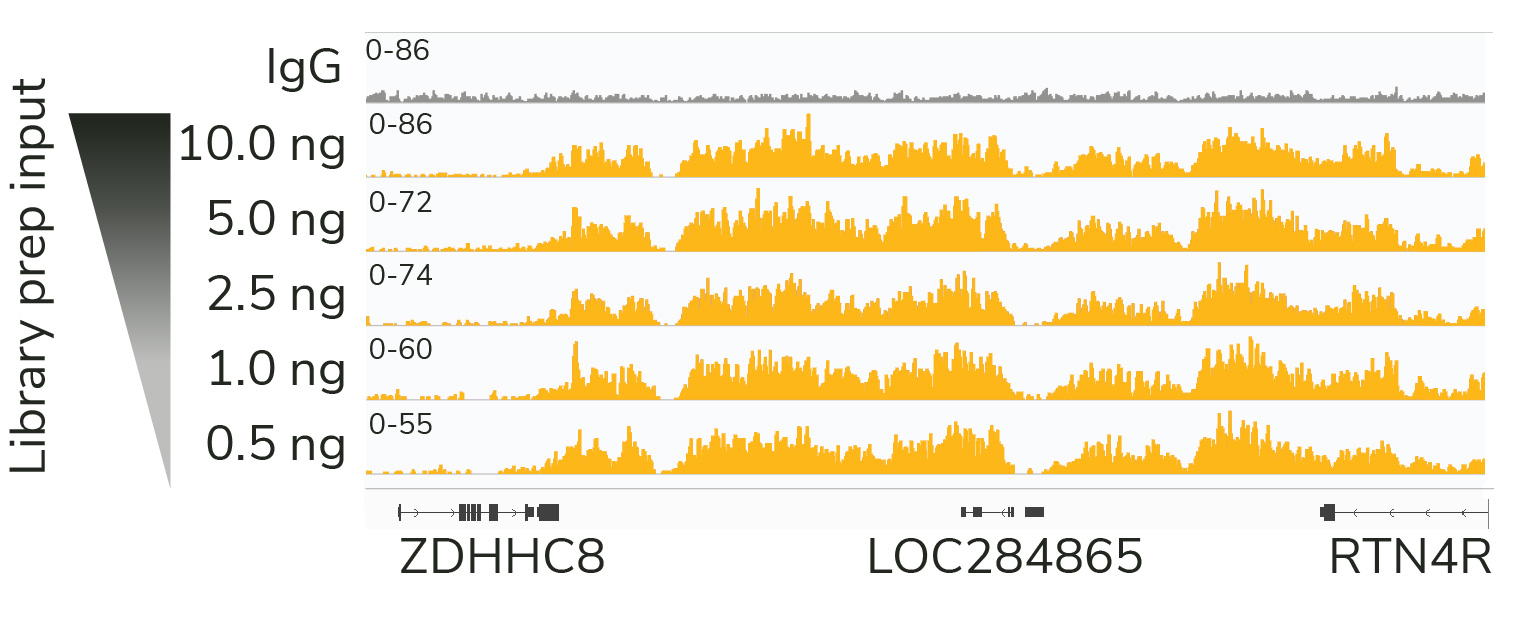
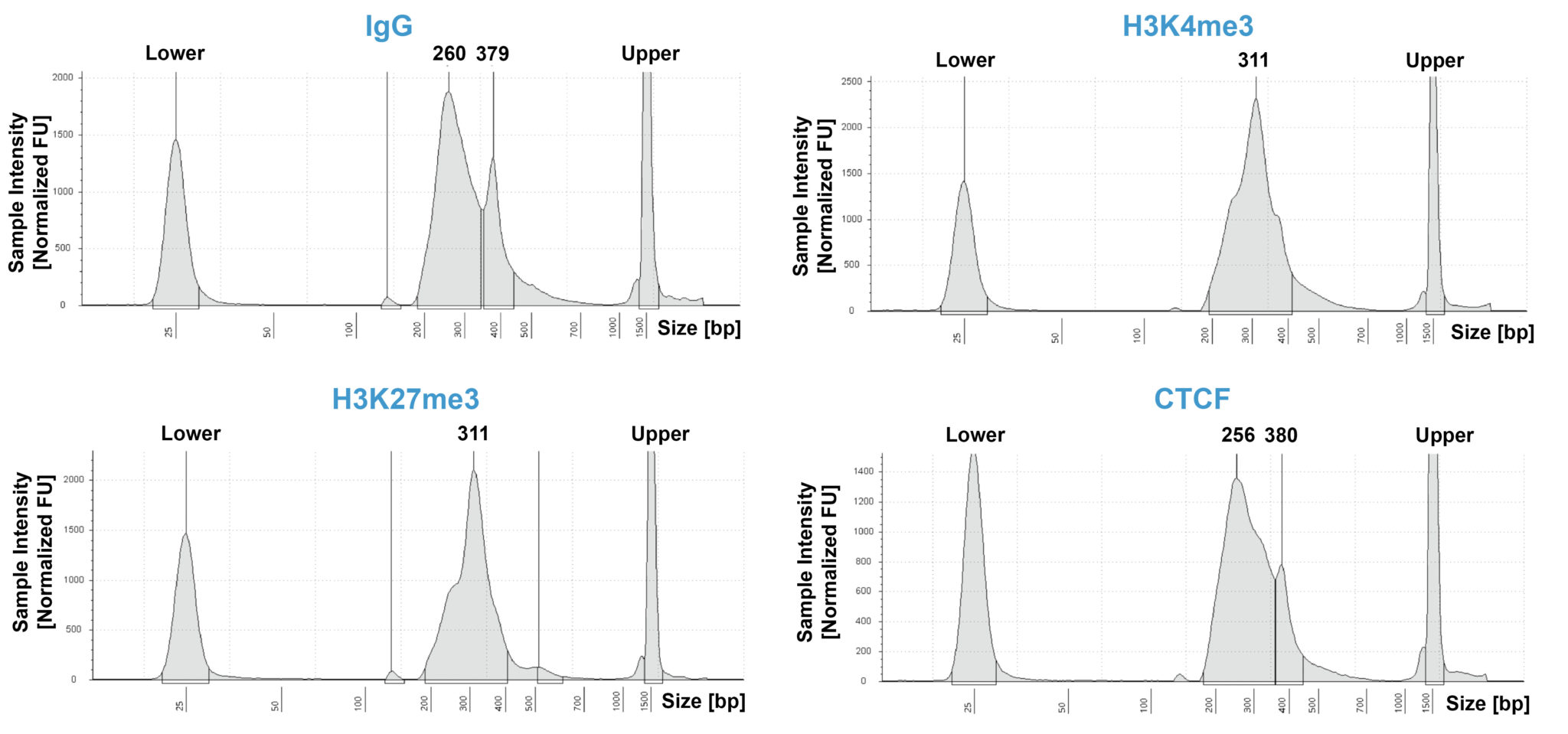
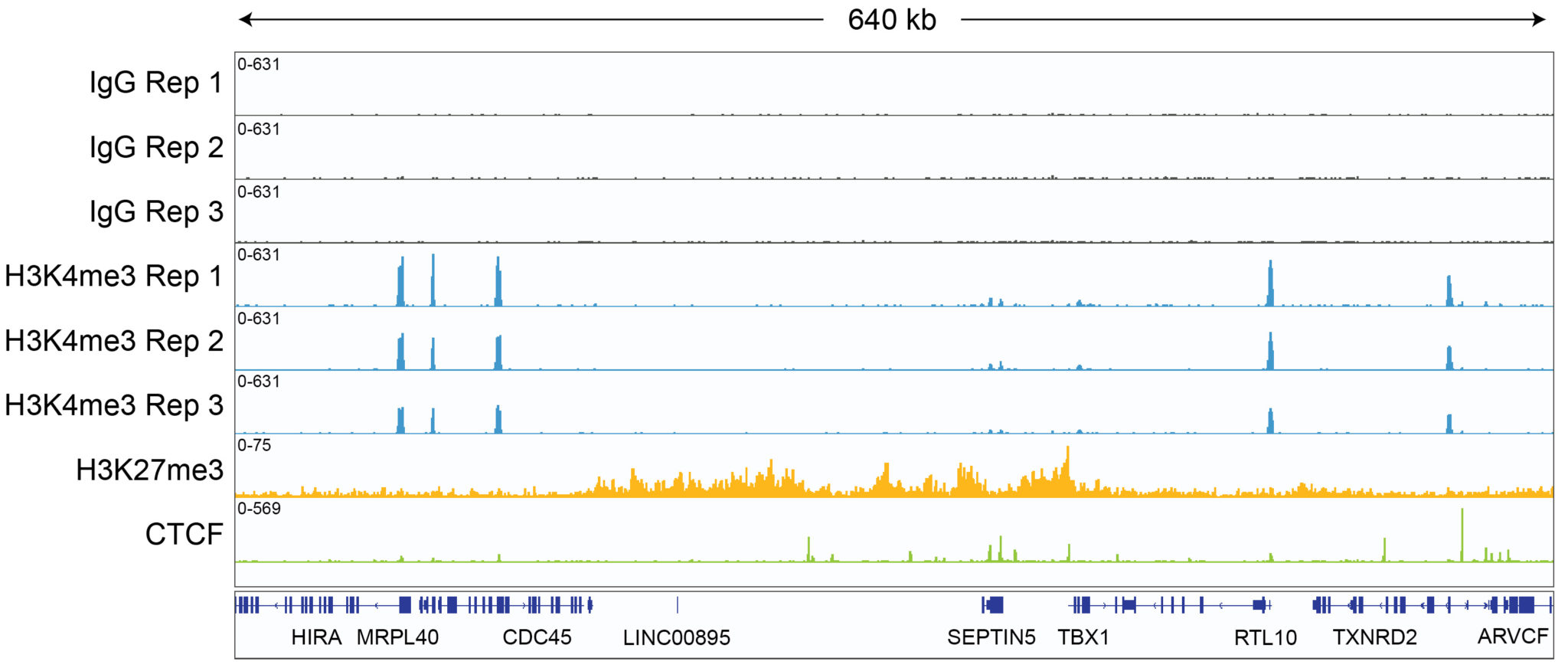
CUT&RUN sequencing libraries described in Figure 2 were sequenced on an Illumina® NextSeq 2000 with a P3 cartridge (paired-end 2×50 cycle). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and blacklist regions. A representative 640 kb window at the SEPTIN5 gene is shown for three replicates (“Rep”) of IgG and H3K4me3 antibodies, as well as individual tracks for H3K27me3 and the transcription factor CTCF, demonstrating the robustness and reproducibility of the workflow with a variety of targets. Sequencing libraries prepared with the CUTANA CUT&RUN Library Prep kit produced the expected genomic distribution for each target. Sample sequencing depth was as follows (millions of reads): IgG Rep 1 (20.1), IgG Rep 2 (16.6), IgG Rep 3 (16.1), H3K4me3 Rep 1 (7.3), H3K4me3 Rep 2 (17.2), H3K4me3 Rep 3 (13.8), H3K27me3 (13.4), CTCF (16.2). Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute).