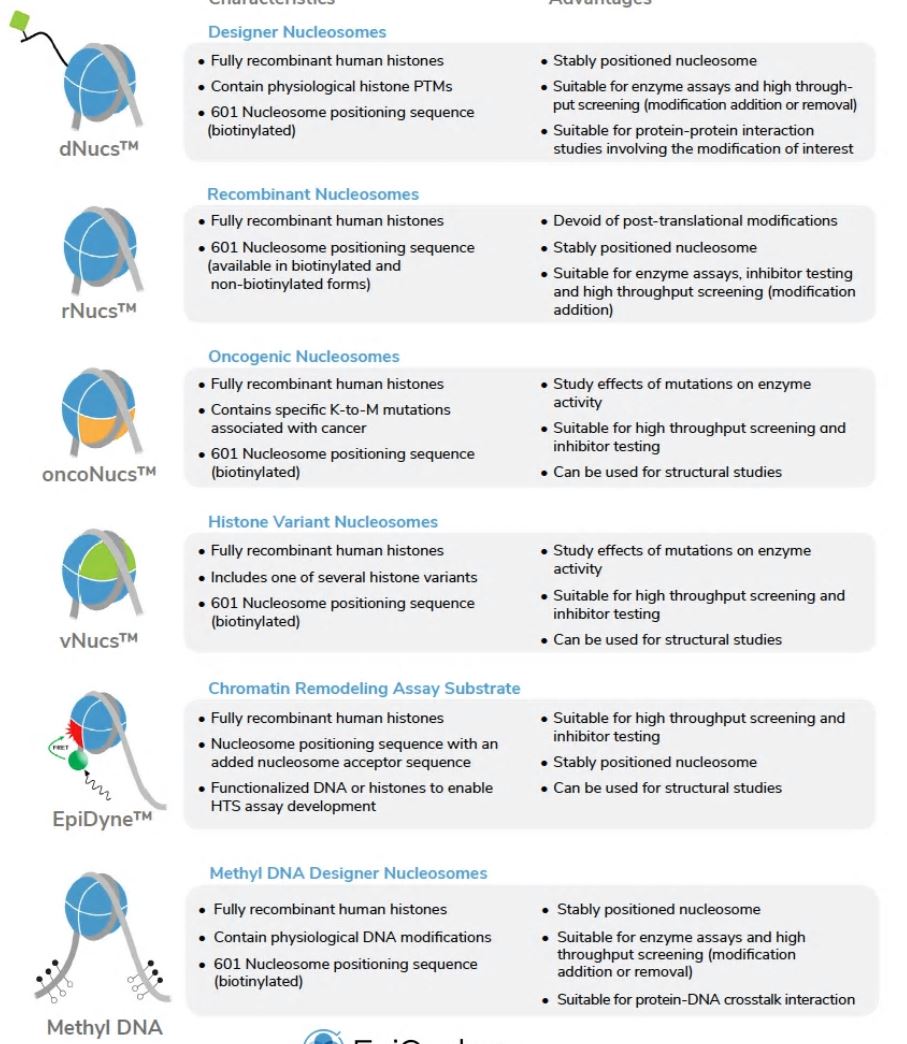
Nucleosome substrates are emerging tools for the in vitro study of chromatin epigenetic regulation, with applications from basic chromatin biology research to high-throughput drug screening. The advent of recombinant designer nucleosomes (dNucs) carrying fully defined covalent modifications of the histone proteins (e.g. lysine acylation) or wrapping DNA (e.g. 5’ methylcytosine) enables novel or dramatically improved approaches (e.g. reader binding, enzymatic assays, antibody profiling) for next generation chromatin research.
In this post we are going to talk about the minimal thresholds for the assembly and quality validation of these reagents.
Recombinant nucleosomes as physiological substrates for drug discovery and drug development
Here at EpyCypher, we have pioneered the commercial development of recombinant nucleosomes for next generation epigenetic assays and tool development (see Table 1).
Mounting evidence demonstrates that nucleosomes are the optimal substrates to characterize many chromatin regulators [1-4]. For example, NSD2 methyltransferase (associated with oncogenic reprogramming in multiple myeloma [5, 6]) requires nucleosomal substrates for in vitro activity [7].
In the following table, there is a list of all the recombinant nucleosomes available from EpiCypher.