Despite the overwhelming popularity of chromatin immunoprecipitation (ChIP) in epigenetics research, ChIP methods have poor yields, high background, and low accuracy / reliability. Although exogenous Drosophila xeno-chromatin has been implemented as a spike-in control for sample normalization1, 2, this natural product is poorly defined as a reagent, making it highly variable between preparations and experiments. Such standards are also not capable of antibody specificity testing or examining technical variation, leaving a large and unmet need for quantitative controls in ChIP experiments.
SNAP-ChIP™ Spike-In Controls are Essential for ChIP Experiments
SNAP-ChIP Spike-Ins Improve Quality of ChIP
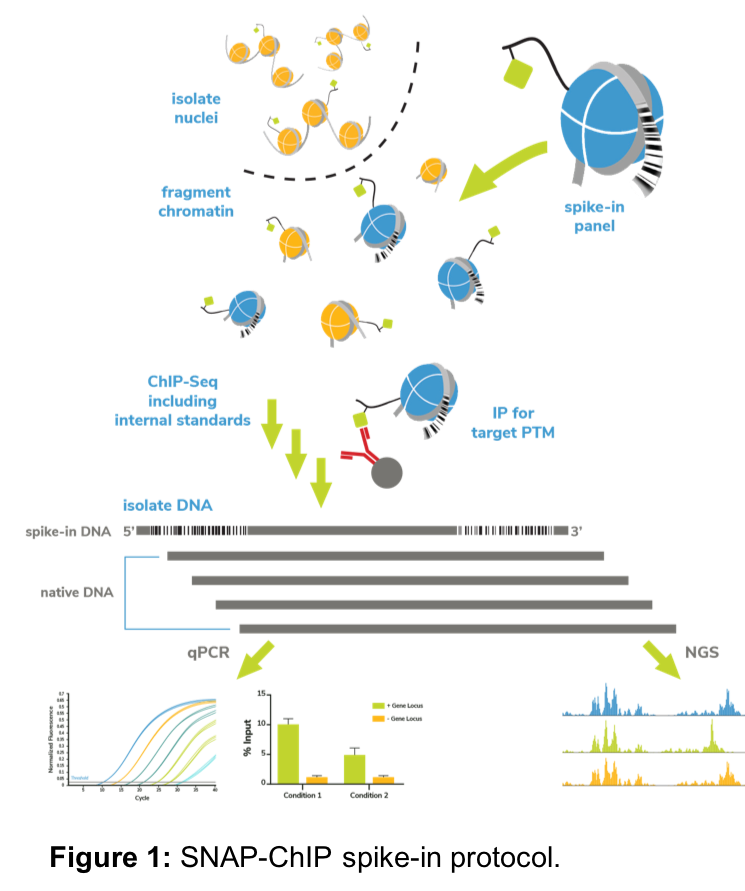
EpiCypher has pioneered the development of recombinant designer nucleosomes (dNucs) for chromatin applications, with an early focus on antibody specificity. Our recently launched SNAP-ChIP™ products and services use panels of DNA-barcoded modified designer nucleosomes (dNucs) as spike-in controls for ChIP experiments (Figure 1). Following immunoprecipitation, we perform qPCR or next-generation sequencing (NGS) to detect PTM-specific DNA barcodes, which quantifies the amount of on- and off-target antibody binding3, 4.
To address growing market needs, EpiCypher has begun leveraging SNAP-ChIP technology to develop best-in-class SNAP-ChIP validated antibodies. In addition, we encourage researchers to perform SNAP-ChIP on their own in-house antibodies, which will confirm and provide renewed confidence in their resulting data.
SNAP-ChIP Spike-Ins Controls: Required for Every Experiment?
We hear this question quite often from customers, who want to know if they need to include SNAP-ChIP spike-in controls in their study design if they are already using a SNAP-ChIP validated antibody. The short answer is, yes; but the explanation requires a more thoughtful demonstration of the myriad advantages of SNAP-ChIP spike-ins.
EpiCypher SNAP-ChIP spike-ins, combined with the use of qPCR primers and probes, are an invaluable tool in several aspects of ChIP methodology, including:
1) Monitoring technical variability between experiments / scientists
2) Assessing lot-to-lot variation in antibody specificity
Both of these approaches will help optimize your ChIP experiments and improve the reliability and accuracy of your data.
STOP/GO Strategy for ChIP-seq
We’ve all been there: You performed your ChIP-seq experiment with three (or more) biological replicates, and everything looks perfect … except that one sample (Figure 2A). That one sample looks drastically different from the rest. What happened? How could this have been avoided?
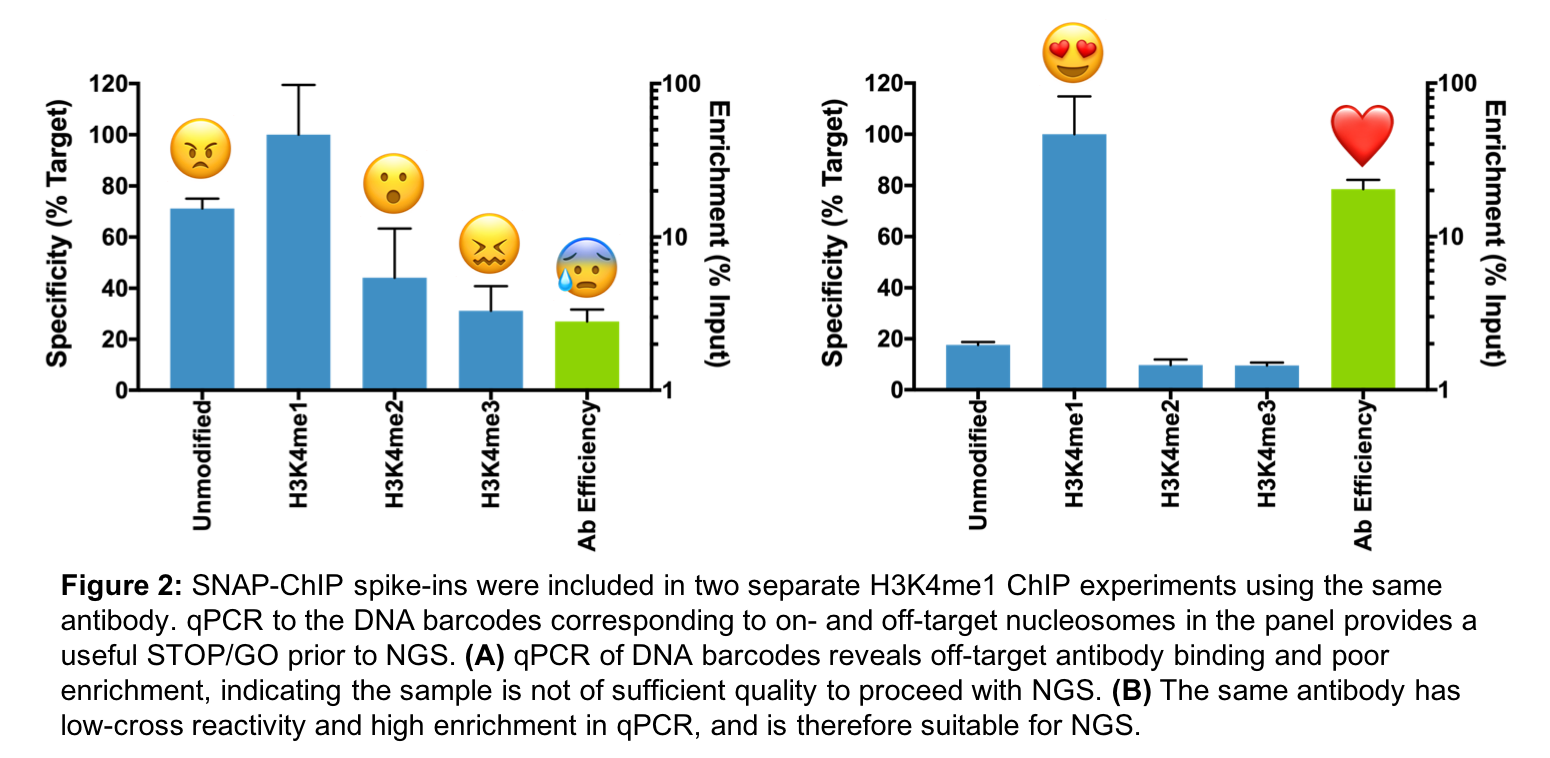
Because SNAP-ChIP panels are added prior to immunoprecipitation (IP), they can be used to monitor unexpected variability between samples. We have developed SNAP-ChIP spike-ins into a useful STOP/GO strategy for ChIP-seq, in which we perform qPCR for dNuc DNA-barcodes following IP, prior to library preparation and NGS (Figure 1).
As shown in Figure 2A vs. 2B, two independent experiments with the same highly specific antibody can still yield drastically different outcomes, ranging from nonspecific binding to reduced IP efficiency. Using our SNAP-ChIP qPCR primers and probes, we have saved considerable time (and money) by identifying poorly prepared samples prior to NGS.
Our strategy fits into a typical ChIP-seq workflow, in which scientists confirm ChIP performance via qPCR for genomic loci previously demonstrated to be enriched for the PTM target, compared to negative controls (non-enriched loci). However, this traditional approach does not control for antibody performance (i.e. antibody specificity and efficiency). Furthermore, if the original genomic loci standards were identified using cross-reactive antibodies, this method is potentially erroneous, and at best problematic3. By providing a highly validated pool of recombinant nucleosomes harboring on- and off-target PTMs, SNAP-ChIP provides defined positive and negative spike-in controls to confidently monitor on-target recovery after ChIP.
Lot-to-Lot Variation in Antibody Specificity
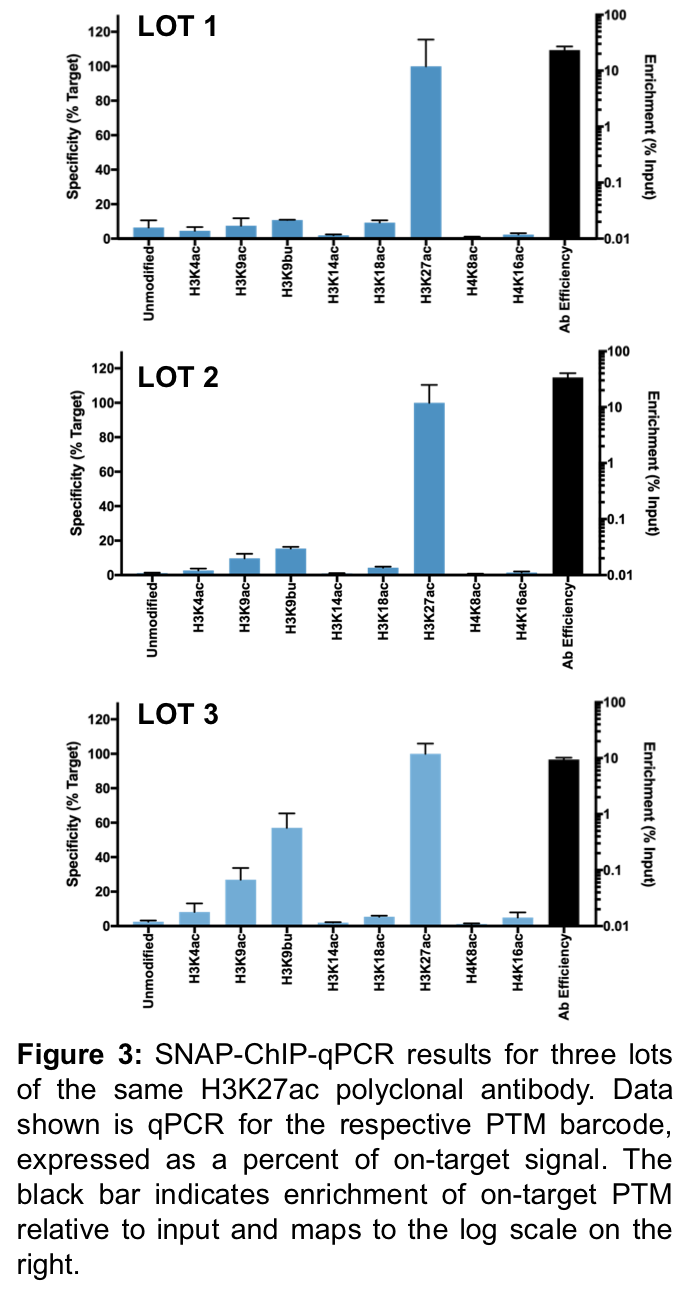
Once you find a SNAP-ChIP certified antibody for your favorite histone PTM, you should be all set, right? Not so fast…
To evaluate the consistency of antibody performance, EpiCypher has tested multiple lots of commercially available ChIP antibodies. Antibody performance (specificity and target enrichment) has been determined for over 200 antibodies tested to date (see Shah et al., 2018 and inquire for unpublished results at info@epicypher.com). Here, we show SNAP-ChIP results for three different lots of a polyclonal antibody (Figure 3).
Unfortunately, lot 3 (Figure 3C) displayed a striking amount of variation from lots 1 and 2 (Figures 3A-B), with significant off-target binding to H3K9ac and H3K9bu, and <10% binding efficiency. After thorough lot-testing of ChIP-grade antibodies, EpiCypher has found that this variability is a common problem, especially for polyclonal antibodies.
Thus, each time you purchase a new lot / preparation of antibody, you should always test it in SNAP-ChIP prior to utilizing the antibody for ChIP experiments.
Quantitative sample normalization
Our early efforts in developing SNAP-ChIP technology focused on defining histone PTM antibody specificity3 and technical monitoring of ChIP experiments (shown above). However, there are other applications for this dynamic platform, including quantitative normalization of ChIP experiments3, 4. EpiCypher is currently working to create more robust and streamlined SNAP-ChIP workflows for ChIP normalization. Testing of these protocols is underway…stay tuned for the next blog post on this topic!
REFERENCES
1. Orlando DA, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep, 2014. 9(3): p. 1163-70. (PubMed PMID: 25437568)
2. Egan B, et al. An Alternative Approach to ChIP-Seq Normalization Enables Detection of Genome-Wide Changes in Histone H3 Lysine 27 Trimethylation upon EZH2 Inhibition. PLoS One, 2016. 11(11): p. e0166438. (PubMed PMID: 27875550)
3. Shah RN, et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell, 2018. 72(1): p. 162-77 e7. (PubMed PMID: 30244833) (PMC6173622)
4. Grzybowski AT, et al. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell, 2015. 58(5): p. 886-99. (PubMed PMID: 26004229) (PMC4458216)