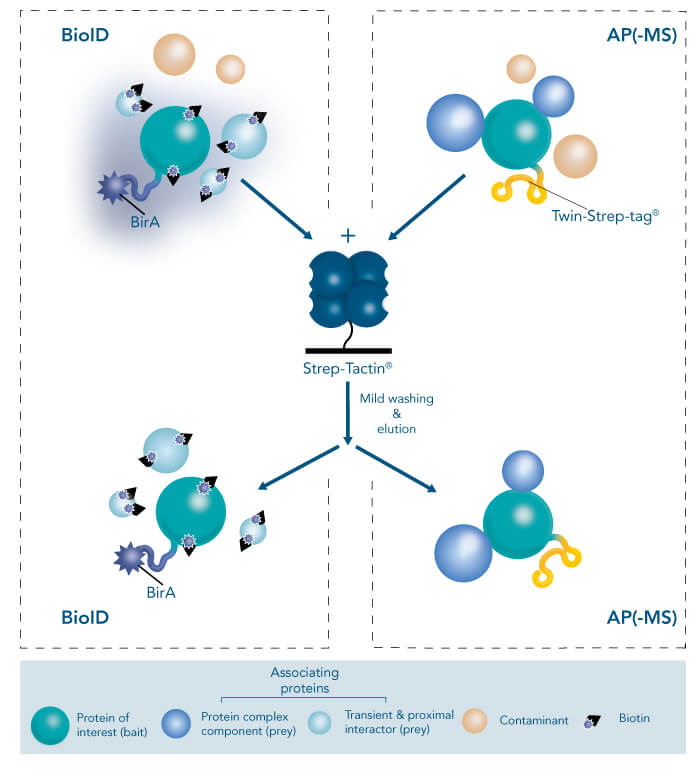
BioID
Products for BioID
Originally the Strep-Tactin®, an engineered streptavidin, was developed for efficient but reversible binding of a peptide that could be used for protein purification – the Strep-tag®. This Strep-tag®/Strep-Tactin® system is now widely used in the field of protein purification.
Nevertheless, the Strep-Tactin® still has the capability to bind biotin, which is used as specific competitor for elution of strep-tagged proteins. The binding affinity of biotin to Strep-Tactin® is not as high as for streptavidin and therefore biotinylated molecules can bind to Strep-Tactin® in a reversible manner and the release from binding is carried out under mild and physiological conditions by adding an excess of free biotin.
This reversible binding of Strep-Tactin® is applicable for purification, detection and immobilization of biotinylated molecules.
Thus, it offers many applications in characterization of protein complexes and mapping of protein-protein interactions using affinity purification-mass spectrometry (AP-MS) or proximity-dependent biotin identification (BioID). While AP-MS techniques rest on the expression of an endogenous protein decorated with high performance epitope tags, such as the Twin-Strep-tag®, and subsequent purification together with associating proteins, BioID involves the expression of a promiscuous biotin ligase BirA fused to the protein of interest, leading to biotinylation of lysine residues of nearby interactors, which can be easily enriched with the aid of Strep-Tactin®. AP-MS provides many advantageous features, including its expediency for both large scale and high-throughput studies as well as quantitative proteomic approaches, whereas BioID allows the capture of even weak or only transient interactions. Consequently, combining both methods results in a comprehensive platform for protein complex and interaction analyses.
Protocol
This step-by-step protocol provides the complete workflow for purification of biotinylated molecules starting with the sample preparation.The procedure is described for purification with both gravity flow columns and for batch purification with magentic beads. The protocol also includes the list of all necessary materials, as well as recommended buffers for washing and elution.
Application Note
Specific Elution and Avoidance of Contaminations in BioID
References
An integrated strategy for mapping the cellular interactome
As laid out in an interesting study in 2018, Liu and colleagues ventured on this project and generated a Gateway® cloning-compatible “multiple approaches combined” (MAC)-tag, which amalgamates the efficient single step Twin-Strep-tag® purification and BioID analysis within a single construct. Conveniently, the utilization of a single affinity reagent drastically reduces the number of required cell lines and improves data reproducibility. In addition, the construct is not only perfectly compatible with downstream processes, including immunohistochemistry and cross-linking mass spectrometry, but further allows deduction of relative spatial distances of proteins within a complex. After examination of the proper localization of 18 MAC-tag fused cellular marker proteins by fluorescence microscopy, the authors took advantage of the high yields and purity as well as the mild native elution of both target proteins and interactors promoted by application of Strep-Tactin®. The localization markers and their 1911 interaction partners identified were used to build a molecular context repository, which functions as reference for “mass spectrometry microscopy” analysis of proteins of interest.
To read more about the integrated workflow and the obtained detailed proteome map, please find the publication here.
A potential redox-regulated switch in the protein import and folding machinery of mitochondrial matrix proteins
Konovalova, S., Liu, X.. Manjunath, P. et al. Redox regulation of GRPEL2 nucleotide exchange factor for mitochondrial HSP70 chaperone. Redox Biol 19 (2018).
To gain insights into the potential roles of vertebrate nucleotide exchange factors of mtHSP70 involved in the translocation of proteins into the mitochondrial matrix, Konovalova et al. exploited BioID proximity labeling and enrichment of the biotinylated proteins using Strep-Tactin® beads to identify potential binding partners of GREPL1 and 2. Based on liquid chromatography-mass spectrometry analysis of the purified protein complexes, they developed a model where the GREPLs form homodimers rather than a hetero-oligomeric subcomplex and GREPL1 is the preferred interactor of mtHSP70 under normal culture conditions. By applying different types of polyacrylamide gel electrophoresis and hydrogen peroxide treatment of HEK293 cells, the researchers revealed that the dimerization of GREPL2 – but not of GREPL1 – is introduced by intramolecular disulfide bond formation. They were able to identify the redox-regulated residue and propose that the protein protects mitochondrial proteostasis by sensing oxidative stress.
For detailed information read the full paper here.
A scaffold protein of eukaryotic ribosomes links signal transduction to both translational efficiency and fundamental nuclear processes
Opitz, N., Schmitt, K., Hofer-Pretz, V. et al. Capturing the Asc1p/RACK1 microenvironment at the head region of the 40S ribosome with quantitative BioID in yeast. MCP (2017). mcp.M116.066654
As first described application of BioID in yeast, Opitz and colleagues employed the labeling technique to characterize the microenvironment of wildtype and mutant Asc1, the S. cerevisiae ortholog of human RACK1 and scaffold protein located at the head of the 40S ribosomal subunit. Besides successfully capturing associated proteins with the aid of Strep-Tactin® Sepharose visualized by streptavidin coupled to horseradish peroxidase, the researchers performed stable isotope labeling with amino acids in cell culture (SILAC) coupled to mass spectrometry for quantification in exponentially growing cells as well as in response to stress. In brief, 40 candidates in the Asc1-vicinity were identified, including translational and transcriptional regulators as well as mRNA-binding proteins, and among them so far undetected physical interactors and additional proximal proteins. Aside from new insights on an established mutant, they report a rearrangement of ribosomes under glucose deprivation and demonstrate the utility of proximity labeling experiments for yeast scaffold proteins.
For detailed information read the full paper here.
A heme-binding protein as possible coordinator of sphingolipid and cholesterol metabolism
Hardt, R., Winter, D., Gieselmann, V. & Eckhardt, M. Identification of progesterone receptor membrane component-1 as an interaction partner and possible regulator of fatty acid 2-hydroxylase. Biochem J 457 (2018).
A different but equally promising combination was presented by Hardt et al. in their aspiration to detect functionally relevant physical interactors of the mammalian sphingolipid synthesis enzyme fatty acid 2-hydroxylase (FA2H), the deficiency of which leads to fatty acid hydroxylase-associated neurodegeneration. In order to reveal and quantify even weak interactions, the researchers combined each of the complementary approaches, BioID labeling and formaldehyde crosslinking, with SILAC and tandem mass-spectrometry. In doing so, they exploited the seamless transition from affinity purification to analytical applications provided by the Strep-tag® technology toolbox: highly efficient purification of their target fused to a Twin-Strep-tag® and crosslinked to the interactors using Strep-Tactin® Macroprep® and detection via Strep-Tactin® HRP conjugate. The possible interactors identified were subsequently investigated by in vivo bimolecular fluorescence complementation, showing a physical and functional interaction of FA2H and PGRMC1, the membrane-associated progesterone receptor component 1, further supported by inhibitor studies. Coherent with already known interdependencies between sphingolipids and sterols, this interaction hints towards a putative regulation point of both synthesis pathways.
For detailed information read the full paper here.
Products for BioID studies and purification of biotinylated proteins
50% suspension to purify Strep-tag®II or Twin-Strep-tag® fusion proteins
Gravity flow column to purify Strep-tag®II or Twin-Strep-tag® fusion proteins

50% suspension to purify Strep-tag®II or Twin-Strep-tag® fusion proteins
Reversible immobilization of biotinylated analytes or Strep-tag®II and Twin-Strep-tag® fusion proteins