| Products & Ordering | |
| 5-Methyl-dCTP NU-1125 mdCTP, 5-mdCTP | 5-Hydroxymethyl-dCTP NU-932 hmdCTP, 5hmdCTP |
| 5-Aza-dCTP NU-1118 | 5-Hydroxymethyl-dC N-1070 hmdC, 5-Hydroxymethyl-2′-deoxycytidine |
| 5-Formyl-dC N-1069 fdC, 5-Formyl-2′-deoxycytidine | 5-Carboxy-dC N-1067 cadC, 5-Carboxy-2′-deoxycytidine |
| N6-Methyl-dATP NU-949 |
Epigenetics
DNA (De)Methylation
5-hydroxymethylcytosine (5-hmC) is a DNA modification that is – along with its analog 5-methylcytosine (5-mC) – an important epigenetic marker associated with critical cellular functions such as stem cell differentiation and brain development[1]. A specific detection of 5-hmC however, is not trivial since traditional sodium bisulfite sequencing does not distinguish between 5-mC and 5-hmC sites[2].
Song et al.[3] reported an approach for the selective detection of 5-hmC residues in genomic DNA based on UDP-6-azide-glucose (UDP-6-N3-Glc) (Fig. 1). T4 Phage β-glucosyltransferase selectively transfers an azide-modified glucose moiety to the hydroxyl group of 5-hmC allowing detection via Copper-free strain-promoted alkyne-azide click chemistry (SPAAC) by
- introduction of a (Desthio)Biotin group (DBCO-containing (Desthio)Biotin → Click Chemistry) for subsequent purification tasks [1,2] or
- introduction of a fluorescent group (DBCO-containing Fluorescent Dyes → Click Chemistry) for subsequent microscopic imaging [1-3].
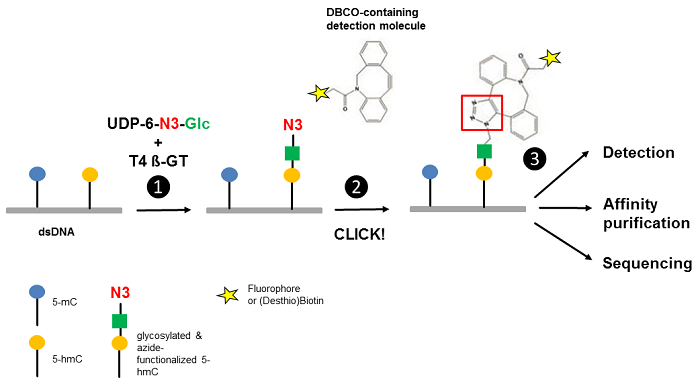 Figure 1 Workflow of 5-hmC detection. Step 1: T4 Phage ß-glycosyltransferase (T4 ß-GT)-mediated glycosylation and simultaneous azide-functionalization of 5-hmC residues using UDP-6-azide-glucose (UDP-6-N3-Glc). Step 2: Copper-free labeling of Azide (N3) with DBCO-containing detection molecule (DBCO-containing (Desthio)Biotin → Click Chemistry or DBCO-containing Fluorescent Dyes → Click Chemistry) forming a stable triazole moiety. Step 3: Detection, affinity purification or sequencing of 5-hmC containing DNA (modified according to [3]).
Figure 1 Workflow of 5-hmC detection. Step 1: T4 Phage ß-glycosyltransferase (T4 ß-GT)-mediated glycosylation and simultaneous azide-functionalization of 5-hmC residues using UDP-6-azide-glucose (UDP-6-N3-Glc). Step 2: Copper-free labeling of Azide (N3) with DBCO-containing detection molecule (DBCO-containing (Desthio)Biotin → Click Chemistry or DBCO-containing Fluorescent Dyes → Click Chemistry) forming a stable triazole moiety. Step 3: Detection, affinity purification or sequencing of 5-hmC containing DNA (modified according to [3]).
| Products & Ordering |
| UDP-6-azide-glucose CLK-076 |
Selected References
[1] Branco et al. (2012) Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature Reviews Genetics 13:7.
[2] Huang et al. (2010) The behaviour of 5-hydroxymethycytosine in bisulfite sequencing. PLOS One 5(1):e8888.
[3] Song et al. (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotech 29(1):68.
[4] Li et al. (2012) Selective Capture of 5-hydroxymethylcytosine from Genomic DNA. J. Vis. Exp. 68:e4441.
[5] Song et al. (2016) Simultaneous single-molecule epigenetic imaging of DNA methylation and hydroxymethylation. PNAS 113(16):4339.
An easy and cost-efficient method for the preparation of 5-(hydroxy)methylated DNA fragments relies on 5-hydroxmethyl-dCTP (5-hmdCTP) or 5-methyl-dCTP (5-mdCTP) that is enzymatically incorporated into DNA with Taq polymerase instead of its natural counterpart dCTP.
5-(hydrox)ymethylated DNA fragments can subsequently be used as sequencing control [1-5] or for pull-down of 5-hydroxymethyl-binding proteins from cellular lysate[6].
Selected references
[1] Booth et al. (2014) Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem. 6 (5):435.
[2] Booth et al. (2013) Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 8 (10):1841.
[3] Yu et al. (2012) Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat. Protoc. 7 (12):2159.
[4] Szwagierczak et al. (2011) Characterization of PvuRts1I endonuclease as a tool to investigate genomic 5-hydroxymethylcytosine. Nucl. Acids Res. 39(12):5149.
[5] Szwagierczak et al. (2010) Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucl. Acids Res. 38(19):e181.
[6] Lafaye et al. (2014) DNA binding of the p21 repressor ZBTB2 is inhibited by cytosine hydroxymethylation. Biochem. Biophys. Res. Commun. 446:341.
