Q1: During construction of Leishmania expression plasmid in E. coli I encountered structural plasmid instability. How can I avoid this problem?
A1: LEXSY expression vectors contain repetitive and homopolymeric DNA stretches within the non-translated regions (utr). Therefore, it is recommended to choose recipient E. coli strains usually tolerating these motives such as XL-10, Stbl2 or Stbl4, DH5α, SURE or related and to incubate them at 30°C (see LEXSY manuals chapter 4.2). If the particular vector/insert combination with your target gene is not tolerated in one of these strains (indicated by plasmid deletions) try another recipient mentioned, rather than analyzing further clones or to repeat transformation with the same recipient strain.
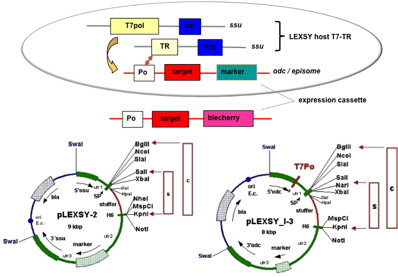
Q2: Can the target gene be introduced into L. tarentolae on a circular plasmid?
A2: Yes, circular plasmids will be maintained in L. tarentolae cells extrachromosomally. For high-yield protein expression from episomal constructs we developed the 4th generation of inducible LEXSY (EGE-1420). However, if integrative expression plasmids are transfected in circular form, the yields of expressed proteins are significantly reduced (to 20-40%) compared to chromosomal integration of a single copy of this target gene. Therefore, it is important to completely linearize the integrative expression plasmids or to transfect the cells with gel-isolated fragments (see LEXSY manuals chapter 4.3).
Q3: There are four different antibiotic resistance markers available for the constitutive expression system. Which marker marker should I prefer?
A3: We recommend to get started with Nourseothricin (NTC) (Nourseothricin) selection using the sat vectors, since establishing recombinant strains is most straightforward with this antibiotic. The order of selection antibiotics regarding their efficiency of eliminating non-recombinant background is: Nourseothricin (NTC) > LEXSY Hygro > LEXSY Bleo > LEXSY Neo.
Q4: Is it possible to co-express two secreted proteins? Would this involve inserting multiple cDNAs into a single expression plasmid or using multiple plasmids with different antibiotic selection markers?
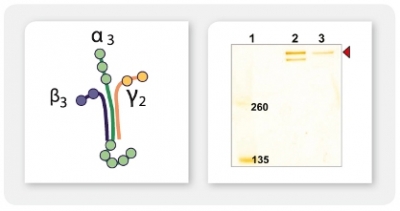
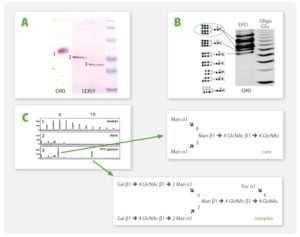
A4: It is possible to co-express up to four secreted or intracellular proteins in the same cell since we offer expression vectors with four alternative antibiotic selection markers and the ssu integration site is present in multiple copies per genome whereupon inactivation of up to four copies does not influence host physiology. Co-expression of up to four gene copies of cytosolic EGFP, superoxide dismutase (SOD1) or secretory erythropoietin (EPO) as well as of a secretory laminin heterotrimer were demonstrated successfully (Jena Bioscience, not published; Gazdag et al. 2010; Phan et al. 2009). Please, refer also to A3 & A8.
Co-expression of multiple target genes from the same vector requires introduction of functional intergenic regions in between the ORFs for proper post-transcriptional processing. This alternative approach was successfully demonstrated for over-expression of human tissue plasminogen activator t-PA (Hemayatkar et al. 2010).
Q5: Can Leishmania be transfected by methods other than electroporation?
A5: Except electroporation, only particle bombardment was described for transfection of Leishmania (Sbicego et al. 1998). All attempts to use chemical transfection methods were up to date unsuccessful (Jena Bioscience, not published).
Q6: How long does it take to get the recombinant Leishmania strain after transfection?
A6: Polyclonal selection of recombinant cell lines in suspension cultures takes routinely two weeks. Within approximately seven days after electroporation differences between the cells electroporated under the same conditions with your target gene or without DNA (negative control) are becoming visible by microscopy. After this it takes another week following dilution into fresh growth medium with selection antibiotic to discriminate visually between the turbid recombinant culture and the clear negative control. However, under certain circumstances more time may be needed for additional 1:10 dilutions (see LEXSY manual for constitutive expression chapter 5.2).
Clonal selection on BHI agar requires about the same time. Colonies start to appear 5-7 days after plating. Approximately 7-9 days after plating they can be transferred into 24 well format and it takes two more days for expansion into 10 ml cultures for further analysis.
Q7: Are all cells of a stable transfected L. tarentolae line considered to harbor the genomic integration of the transfected expression cassette after selection?
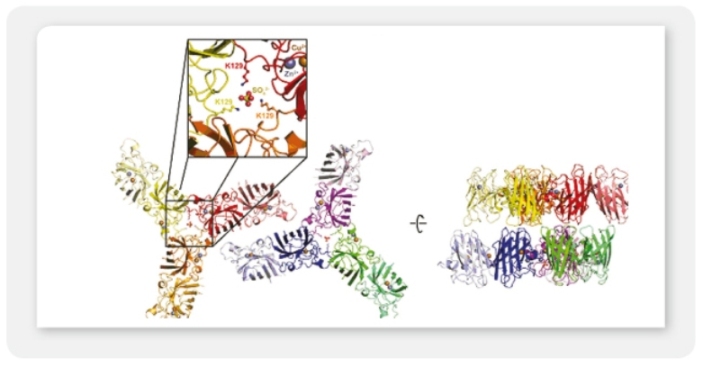
A7: Yes, we showed by fluorescence microscopy, that virtually all cells of a stable LEXSY line selected in suspension culture expressed the transfected egfp gene (see Figure 1 in the LEXSY brochure 2011).
Q8: What are the relative advantages and disadvantages of constitutive versus inducible expression in LEXSY? Will constitutive expression give us high yields?
A8: The features of constitutive and inducible LEXSY configurations are compared in the LEXSY Configurations section of the JBS website and in the LEXSY Brochure 2011 pages 7-10. The inducible LEXSY is recommended for target proteins, which are potentially toxic for the expression host (e. g. proteases, nucleases, kinases). However, this configuration allows co-expression of only two target proteins, whereas with the constitutive LEXSY four target proteins can be co-expressed in the same cell.
Concerning the yields it was found with several cytosolic proteins that the yields in the inducible configuration were 5-10x higher as compared to the constitutive, whereas in case of several secretory proteins the yields were comparable in both configurations.
Q9: Does codon usage bias play a role in LEXSY and do you recommend using a synthetic gene optimized for the Leishmania codon usage? Our target genes are AT rich.
A9: Yes, whenever your budget allows, we recommend gene synthesis with L. tarentolae optimized codon usage because several synthetic genes were expressed at high levels. However, these findings are case sensitive since there are also numerous examples of highly expressed genes with native (e.g. human) codon bias and using synthetic genes does not guarantee higher expression levels than using the corresponding native genes.
Also, codon optimized gene synthesis changes the mRNA sequence and possibly its stability which may contribute to the final target protein yield. In one case we showed, that enhanced mRNA stability was indeed the reason for (or at least significantly contributed to) higher expression levels of a synthetic gene versus the native variant. To answer this question for your particular gene you should analyze expression at mRNA and protein levels.
Q10: What are the optimal conditions for expression of my recombinant target protein in LEXSY?
A10: The optimal conditions for expression of target proteins in LEXSY should be determined individually. Recommended are time course experiments under variable growth conditions such as static or agitated suspension cultures. We found e.g. that secreted proteins could be obtained at higher yields from agitated suspension cultures.
In case of constitutive expression you may also consider, that the ssu locus – the integration site of target genes – is transcribed at highest rates during the growth phase of the cultures. For inducible LEXSY, variation of time point of induction and of inducer concentration may influence target protein yields (see inducible LEXSY manuals chapter 8.6). If the target proteins are stable and non-toxic to the host, induction at inoculation usually results in the best yields.
Q11: Can target proteins be purified with affinity tags from LEXSY?
A11: Yes, affinity purification with His- or Strep-tags worked well in LEXSY with Strep-tags appearing to be more specific than His-tags. The LEXSY vectors are provided with the option for fusion of the target protein sequence to a C’ hexahistidine tag. Other short tag sequences can be added directly to the gene of interest before insertion into the expression vector. This way additional amino acids resulting from vector sequences (polylinker) can be avoided.
Q12: A key problem in protein expression in E. coli is the insolubility of the recombinant proteins. Are the proteins expressed in LEXSY soluble and did you observe inclusion bodies?
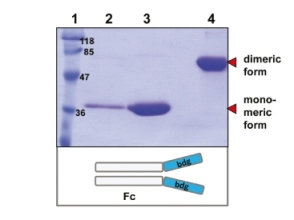
A12: LEXSY has the advantage of a fully functional eukaryotic protein biosynthesis, folding and modification machinery. A eukaryotic – especially mammalian – target protein is expected to fold more natively in Leishmania than in E. coli. All proteins we tested so far were soluble and proteins insoluble in E. coli were expressed in soluble form in LEXSY (for examples see p.18 of the LEXSY brochure 2011 and the chapter “Selected Applications” of the LEXSY section of the Jena Bioscience website). In addition to folding, LEXSY performs mammalian-type posttranslational modifications as N-glycosylation which may be essential for activity, function and stability of the target protein.