GMP-Grade RNA Enzymes
Products
Overview of KACTUS Enzymes for mRNA Synthesis
The key to linear mRNA therapy lies in efficiently synthesizing high-quality mRNA in vitro and then delivering mRNA to the human body through a delivery system. The in vitro synthesis of mRNA cannot be done without high-quality raw material enzymes. KACTUS has a particular focus on the mRNA therapy market we’ve used our unique functional recombinant protein and high-activity enzyme production platform, SAMS, to develop a full series of high-quality raw material enzymes required for in vitro synthesis of mRNA (GMP-Grade & GMP-Ready). Our GMP products have analytical method verification and undergo comprehensive quality release testing to meet the requirements for upstream raw material enzymes.
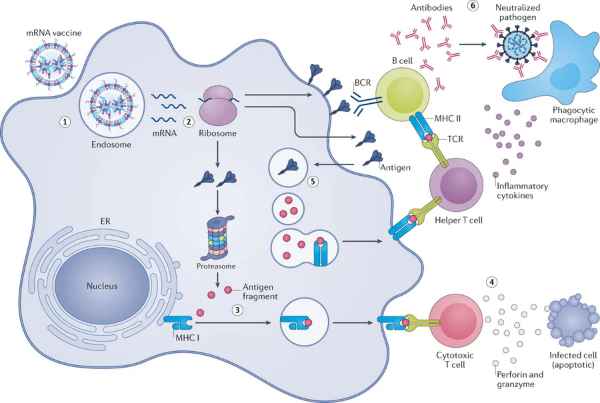
Figure 1. Mechanism of action of mRNA vaccines [1].
Enzymes for every step of mRNA synthesis.
Restriction Endonucleases for Plasmid Linearization
The template for mRNA in vitro transcription is typically plasmid DNA, and needs to be linearized before transcription. Linearization ensures the acquisition of mRNA transcripts of definite length and sequence. KACTUS provides restriction endonucleases such as BsaI, BspQI, SapI, XbaI, etc., for the preparation of linearized templates.
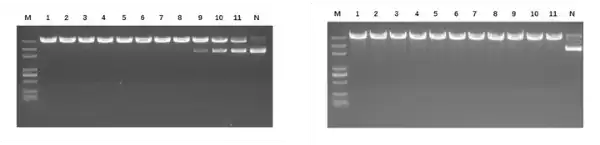
Figure 2. (Left) BsaI enzyme activity detection. 1μg of plasmid was added to a 20 μL system, along with 1 μL BsaI (well 1 is undiluted, wells 1-11 are serial 2-fold dilutions), and digested for 30 minutes. The enzyme activity is ≥20 kU/mL. (Right) BsaI star activity detection. 1 μg of plasmid is added to a 20 μL system, along with 1 μL BsaI (well 1 is undiluted; wells 1-11 are serial 2-fold dilutions) and digested for 16 hours. No star activity is observed.
Products
T7 RNA Polymerase, DNase I, Pyrophosphatase, Murine RNase Inhibitor
In Vitro Transcription (IVT) is a technique that generates mRNA by mimicking the internal transcription process enzymatically, using linearized plasmid DNA as a template. KACTUS provides GMP-Grade T7 RNA Polymerase, Inorganic Pyrophosphatase, Murine RNase Inhibitor, and DNase I to assist in synthesizing high-quality mRNA.
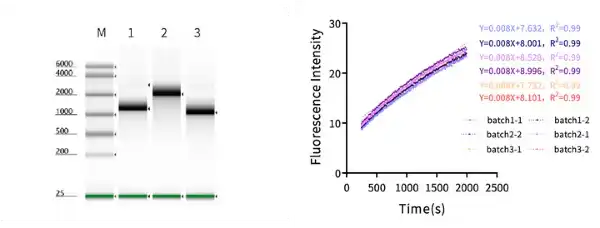
Figure 3. (Left) Detection of T7 RNA Polymerase transcription efficiency. In a 20 μL system, three plasmids were used as templates (1 with poly A tail, 2 and 3 without poly A tail), reacting at 37℃ for 2 hours, with transcription lengths of 2000 nt, 4000 nt, and 2000 nt respectively. T7 RNA Polymerase can efficiently transcribe mRNA of different lengths. (Right) Probe-based detection of T7 RNA Polymerase activity. The detection principle is that when the probe and transcription product bind specifically, a conformational change occurs, causing a change in fluorescence intensity; three batches of T7 RNA Polymerase are tested, each with two experimental replicates, their slopes are basically consistent, indicating the activity of the three batches of T7 RNA Polymerase is stable.
Products
Enzymes for Capping and Tailing Modification
The mRNA produced by IVT has a triphosphate group at the 5’ end, which has strong immunogenicity, necessitating capping and tailing modifications in industrial production. KACTUS offers Vaccinia Capping Enzyme and mRNA Cap 2´-O-Methyltransferase needed for capping modifications, as well as E.coli Poly(A) Polymerase required for tailing modifications. To precisely quantify important indicators such as “capping rate” and “Poly(A) tail product length,” we have carefully established an LC-MS platform and developed mature mass spectrometry detection methods to control the release of enzyme products by batch. We can also provide capping rate and tailing determination services according to your needs.
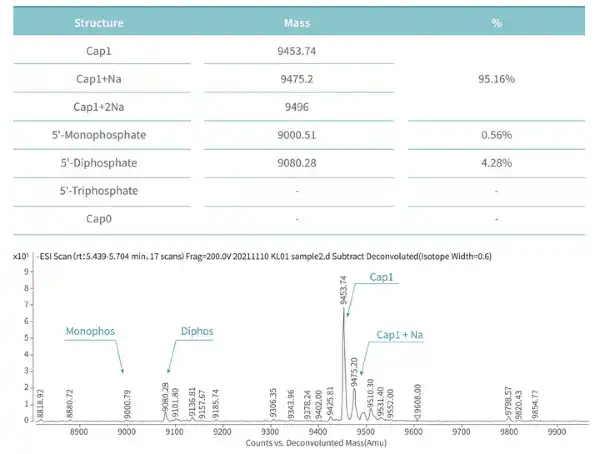
Figure 4. mRNA is capped with Cap1 structure using Vaccinia Capping Enzyme and mRNA Cap 2´-O-Methyltransferase from KACTUS. The capping rate is 95.16% detected via our LC-MS platform. (Note: The capping rate is related to the performance of the enzyme and factors such as the secondary structure of mRNA.)
Products
Enzymes for circRNA In Vitro Synthesis
Linear mRNA obtained from IVT can be further circularized to obtain circRNA. The circularization methods include Type I intron self-splicing, Type II intron self-splicing, T4 RNA Ligase, etc. KACTUS provides T4 RNA Ligase I, T4 RNA Ligase II, and RNase R for in vitro RNA circularization and purification processing of circRNA.
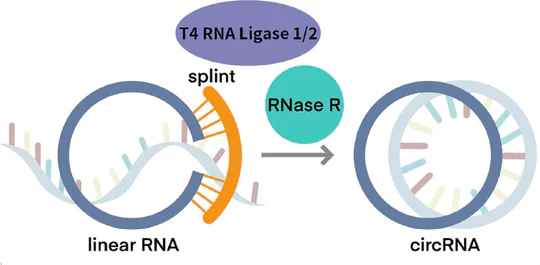
Figure 5. circRNA In Vitro Synthesis.
RNase R, GMP-Ready
KACTUS has developed a GMP-Ready RNase R enzyme to aid in digestion of linear RNA for circRNA development.

Products
Browse All mRNA Enyzmes
GMP-Grade: Currently manufactured according to cGMP guidelines.
GMP-Ready: Industrial-Grade enzyme that can be transitioned to our 100,000 sq ft GMP-Grade facility with GMP documentation and testing.
Drug Master Files: Submitted to the FDA Drug Master Files (DMF)
| Order Now | Type | Product Name | Grade | DMF # (if applicable) |
| GMP-BSA-EE101 | Restriction Endonuclease | BsaI | GMP-Grade | 37503 |
| BSP-BE101 | Restriction Endonuclease | BspQI | GMP-Ready | – |
| KPN-KE101 | Restriction Endonuclease | KpnI | GMP-Ready | – |
| NRU-RE101 | Restriction Endonuclease | NruI | GMP-Ready | – |
| PVU-PE101 | Restriction Endonuclease | PvuII | GMP-Ready | – |
| SAL-SE101 | Restriction Endonuclease | SalI | GMP-Ready | – |
| SAP-SE101 | Restriction Endonuclease | SapI | GMP-Ready | – |
| XBA-EE101 | Restriction Endonuclease | XbaI | GMP-Ready | – |
| GMP-DNI-EE001 | IVT | DNase I | GMP-Grade | 38032 |
| GMP-RNI-ME101 | IVT | Murine RNase Inhibitor | GMP-Grade | 38031 |
| GMP-PYR-YE101 | IVT | Pyrophosphatase, Inorganic | GMP-Grade | 38030 |
| GMP-T7P-EE101 | IVT | T7 RNA Polymerase | GMP-Grade | 37660 |
| GMP-MEH-VE101 | Capping | mRNA Cap 2′-O-Methyltransferase | GMP-Grade | 38029 |
| GMP-VCS-VE101 | Capping | Vaccinia Capping Enzyme | GMP-Grade | 38028 |
| RNR-EE001 | Circular RNA | RNase R | GMP-Ready | – |
| TRL-BE101 | Circular RNA | T4 RNA Ligase I | GMP-Ready | – |
| TRL-BE103 | Circular RNA | T4 RNA Ligase II | GMP-Ready | – |
| PLA-EE101 | Tailing | E.coli Poly (A) Polymerase | GMP-Ready | – |
References
[1] Chaudhary, N., Weissman, D. & Whitehead, K.A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov 20, 817–838 (2021).
Browse GMP-Grade RNA Enzyme Products by Category
GMP-Grade T7 RNA Polymerase
Catalog #GMP-T7P-EE101
FDA Drug Master Files #037660
Designed for commercial-scale mRNA manufacturing.

Product Features
High Yield
Batch-to-Batch Consistency
Low dsRNA

Stability Testing
Inventory in stock

Regulatory Documentation
About T7 RNA Polymerase
T7 RNA Polymerase (T7RNAP) is a protein encoded by phage T7 DNA recombinantly expressed in Escherichia coli. T7RNAP, a DNA-dependent 5’→3′ RNA polymerase, highly specifically recognizes the T7 promoter sequence (5′-TAATACGACTCACTATAG-3′). The template is single-stranded or double-stranded DNA containing the T7 promoter sequence. T7RNAP synthesizes an RNA strand using NTPs, complementary to the DNA template, downstream of the promoter.
About Our Enzyme Engineering & Expression Optimization
We engineered our T7 RNA Polymerase using Structure Aided Multiplex Screening (SAMS), our unique innovative functional recombinant protein engineering platform. It has also undergone optimization of the E. coli expression system, purification process, and buffer formulation. We’ve designed our T7 to produce low error rates, high efficiency, and high processivity. We’ve optimized the enzyme for yield and purity, making it both practical and affordable for large-scale production of mRNA / mRNA synthesis.
About Our GMP Compliance
We manufacture our T7 RNA Polymerase in our cGMP compliant facility. Moreover, we’ve submitted our GMP T7RNAP to the FDA Drug Master Files (DMF #037660). Customizable regulatory support documentation including batch production records, stability testing, CoA, etc. is available.
Customizable GMP Documentation Package:
→ Datasheet
→ CoA
→ CoO
→ MSDS
→ Melamine Statement
→ TSE/BSE Statement
→ Nitrosamine Statement
→ DMF Filing
Product Information
| Parameter | Specification |
| Catalog # | GMP-T7P-EE101 |
| FDA Drug Master Files | #37660 |
| Available Sizes | 1KU / 50KU / 1MU |
| Concentration | 50U/µL |
| Source | Expressed in an E.coli strain that carries the T7 RNA Polymerase gene. |
| Molecular Weight | 98 kD |
| Form | Liquid |
| Storage | Store at -20 ± 5°C, avoid repeated freezing and thawing |
| Unit Definition | One unit is defined as the amount of enzyme required to incorporate 1 nmol ATP into acid-insoluble material in a total reaction volume of 50µL in 1 hour at 37°C. |
| Applications | In vitro transcription (IVT) RNA analysis Radiolabeled RNA probe preparation Hybridization RNase protection assay |
Quality Release Specifications
| Parameter | Specification |
| Activity (Molecular Beacon) | ≥ 50 kU/mL |
| Purity (SEC-HPLC) | ≥ 95% |
| Residual DNase | Negative |
| Residual RNase | Negative |
| Residual Protease | Negative |
| Endotoxin | ≤ 10 EU/mL |
| Residual Host Cell DNA | ≤ 100 pg/mg |
| Residual Host Protein | ≤ 20 ng/mg |
| Residual Heavy Metal | ≤ 10ppm |
| Bioburden | ≤ 1 CFU/10mL |
| Residual Nickel Salt | ≤ 10ppm |
Performance Validation
Overall Performance
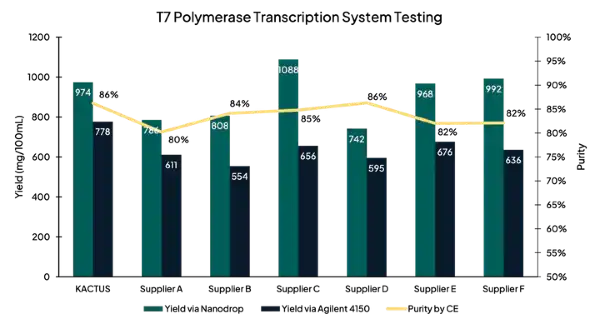
Figure 1. After synthesizing mRNA in an IVT reaction, we assessed RNA transcript yield using nanodrop and Agilent 4150 tapestation. We assessed purity using capillary electrophoresis (CE). Overall, yield and purity of our T7RNAP was comparable to leading suppliers.
Low dsRNA in transcripts
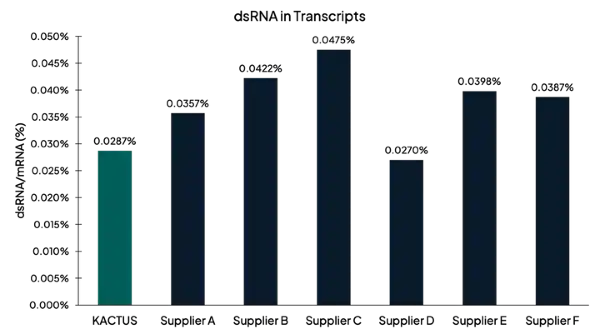
Figure 2. dsRNA measured after in vitro transcription using J2-based ELISA. KACTUS T7 has low dsRNA contamination comparable or superior to leading competitors
High yield for in vitro transcription
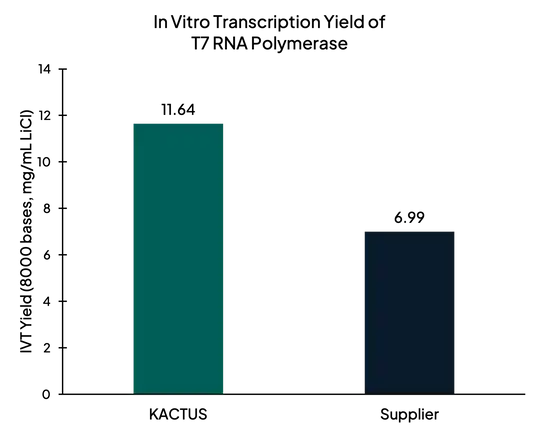
Figure 3. After synthesis of mRNA, in vitro transcription yield of T7 RNA Polymerase measured with Agilent 4150 Tapestation. KACTUS T7 had 65% higher yield than leading supplier.
Consistent activity across batches
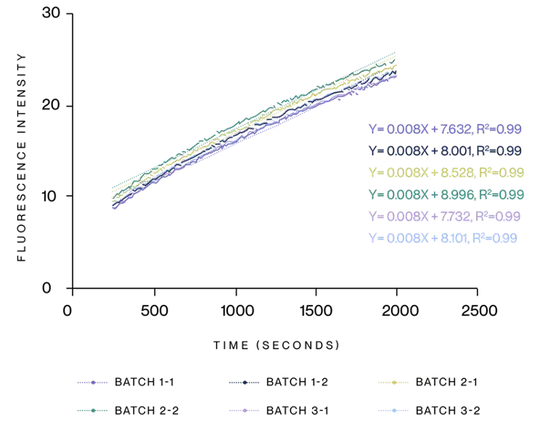
Figure 4. Detection of T7 RNA Polymerase by molecular beacon. We analyzed three batches of T7 RNA Polymerase and found similar reaction kinetics across all batches.
KACTUS offers a selection of GMP-Grade and GMP-Ready Restriction Endonucleases for plasmid linearization prior to in vitro transcription. Our restriction enzymes are expressed in E. coli and have high purity and batch consistency.
GMP-Grade: Manufactured according to cGMP guidelines.
GMP-Ready: Industrial-grade manufacturing transferrable to GMP-level manufacturing.
| Enzyme | Recognition Site | Grade | Catalog # |
| BsaI | 5’…GGTCTC(N)₁↓…3’ 3’…CCAGAG(N)₅↑…5’ | GMP-Grade DMF #037503 | GMP-BSA-EE101 |
| BspQI | 5’…GCTCTTC(N)₁↓…3’ 3’…CGAGAAG(N)₄↑…5’ | GMP-Ready | BSP-BE101 |
| KpnI | 5’…GGTAC↓C…3’ 3’…C↑CATGG…5’ | GMP-Ready | KPN-KE101 |
| NruI | 5’…TCG↓CGA…3’ 3’…AGC↑GCT…5’ | GMP-Ready | NRU-RE101 |
| SalI | 5’…G↓TCGAC…3’ 3’…CAGCT↑G…5’ | GMP-Ready | SAL-SE101 |
| SapI | 5’…GCTCTTC(N)₁↓…3’ 3’…CGAGAAG(N)₄↑…5’ | GMP-Ready | SAP-SE101 |
| XbaI | 5’ …T↓CTAGA… 3’ 3’ …AGATC↑T… 5’ | GMP-Ready | XBA-EE101 |