LSBio’s library of coronavirus products includes numerous kits for the detection of SARS, SARS-CoV-2, MERS, HCoV, FIPV, and other common coronaviruses. We offer COVID-19 real-time quantitative PCR (RT-qPCR) kits for the direct detection of SARS-CoV-2 RNA and a wide range of ELISA kits to detect human IgG/IgM, coronavirus proteins, and host proteins relevant to COVID-19 disease progression.
lifespan Biosciences COVID-19
Research Solutions for Coronavirus – Lifespan Biosciences
Coronavirus Research Products
Viral RNA Detection Kits (RT-qPCR)
Real-Time qPCR (RT-qPCR) is a technique that allows researchers to quantitatively detect the presence of specific RNA, such as SARS-CoV-2, within a sample. Following nucleic acid extraction and reverse transcription of RNA to cDNA, the polymerase chain reaction is used to amplify the target DNA, which is then detected using fluorescently labeled highly-specific probes. As the reaction progresses, the release of fluorescence can be monitored, providing a means of quantifying the target. In response to the COVID-19 pandemic, we offer both Real-Time qPCR kits and sample collection kits. The RT-qPCR kits require access to a real-time quantitative PCR instrument.
This rapid test is for research use only and has not been approved by the FDA for clinical diagnostic use. Samples collected and processed for this test are potentially infectious, and should be handled only by trained laboratory professionals in a BL-2 laboratory or equivalent setting with a laminar flow hood and personal protective equipment.
| Catalogue number | Product name |
| LS-K1073 | SARS-CoV-2 RT-qPCR Detection Kit (Dual Gene ORF1ab/N) |
| LS-K1075 | SARS-CoV-2 RT-qPCR Detection Kit (Dual Gene RdRP/N) |
| LS-K1078 | One-Step Core RT-qPCR Kit (No Primers or Probes) |
| LS-K1080 | SARS-CoV-2 RT-qPCR Detection Kit (Triple Gene RdRP/N/E) |
| LS-K1081 | SARS-CoV-2 RT-qPCR Detection Kit (Dual Gene RdRP/N) |
ELISA Detection Kits

LSBio provides a range of Sandwich ELISA kits for the quantitative detection of numerous coronavirus proteins, including SARS-CoV and SARS-CoV-2 spike and nucleocapsid proteins. We also offer kits which detect the SARS-CoV-2 receptor ACE2, protease TMPRSS2, and numerous inflammatory proteins, such as IL-6, IL-10, and TNF-alpha.
ELISA for SARS-CoV-2 IgG/IgM Antibody Detection
| Catalogue number | Product name |
| LS-F74082 | Human Anti-SARS-CoV-2 N/S1 IgG (Direct ELISA) ELISA Kit |
| LS-F74084 | Human SARS-CoV-2 Spike IgM (Direct ELISA) ELISA Kit |
| LS-F74085 | Human SARS-CoV-2 Spike IgG (Direct ELISA) ELISA Kit |
| LS-F74078 | Human Anti-SARS-CoV-2 IgG (Direct ELISA) ELISA Kit |
| LS-F74079 | Human Anti-SARS-CoV-2 IgM (Direct ELISA) ELISA Kit |
| LS-F74081 | Human Anti-SARS-CoV-2 N IgM (Direct ELISA) ELISA Kit |
| LS-F74080 | Human Anti-SARS-CoV-2 N IgG (Direct ELISA) ELISA Kit |
| LS-F74086 | SARS-CoV-2 Nucleoprotein (Sandwich ELISA) ELISA Kit |
ELISA for SARS-CoV-2 Viral Proteins
| Catalogue number | Product name |
| LS-F74078 | Human Anti-SARS-CoV-2 IgG (Direct ELISA) ELISA Kit |
| LS-F74079 | Human Anti-SARS-CoV-2 IgM (Direct ELISA) ELISA Kit |
| LS-F74081 | Human Anti-SARS-CoV-2 N IgM (Direct ELISA) ELISA Kit |
| LS-F74080 | Human Anti-SARS-CoV-2 N IgG (Direct ELISA) ELISA Kit |
| LS-F74086 | SARS-CoV-2 Nucleoprotein (Sandwich ELISA) ELISA Kit |
LSBio offers antibodies, proteins, and cDNAs specific to coronavirus proteins and host proteins involved in coronavirus disease progression. Reagents for investigating SARS-CoV-2, SARS-CoV, MERS, and HCoV coronavirus structural and non-structural proteins, as well as coronavirus-interacting host proteins such as ACE2, TMPRSS2, Cathepsin, and ADAM17 are available and approved for use in multiple applications. We have a wide selection of related recombinant proteins, native proteins, and over-expression lysates. Furthermore, we provide expression-ready ORF cDNA clones to coronavirus genes of prominent research interest.
Antibodies
LSBio offers over 150 antibodies to structural and non-structural coronavirus proteins, including antibodies for the detection of the spike (S), nucleocapsid (N), and envelope (E) proteins of both SARS-CoV and SARS-CoV-2. We also offer antibodies which detect human proteins known to interact with coronaviruses or to contribute to COVID-19 disease progression. Many of our antibodies are available in multiple conjugated forms and are designed for use in a range of assays including ELISA, IHC, Western Blot, IF, and Flow Cytometry.
Recombinant Proteins
View LSBio’s library of high-quality recombinant coronavirus proteins, produced in a variety of expression systems and ready for use in functional assays, high throughput screens, structural analysis studies, and a variety of other applications. Full length recombinant proteins to SARS-CoV-2 structural proteins are available for your COVID-19 research needs.
Expression-Ready ORF cDNA Clones
LSBio offers expression-ready ORF clones to a range of coronavirus genes intended for immediate transfection. SARS-CoV-2 clones are available for stable or transient mammalian and lentiviral expression.
Coronavirus and SARS-CoV-2 Inhibitors
LSBio offers a range of research-use-only biochemicals that show potential inhibitory action against various coronaviruses such as SARS-CoV-2. These include direct inhibitors of coronavirus spike, nucleocapsid, protease, and polymerase proteins. We also offer various chemicals that block host proteins utilized by coronaviruses for infection and replication.
SARS-CoV-2 Spike (S) Protein Inhibitors
The spike (S) protein is responsible for attachment to and fusion with the host cell. For both SARS-CoV and SARS-CoV-2, the host receptor is known to be angiotensin converting enzyme 2 (ACE2) (Hoffmann, 2020). The spike protein is proteolytically cleaved into two subunits, S1 and S2. The S1 subunit binds to ACE2 presented on the host cell surface, and the S2 subunit is responsible for fusion with the host cell. Target cells include pneumocytes and macrophages expressing ACE2 in the lung, as well as ACE2-positive epithelial cells in the lung, gastrointestinal tract, and liver (cholangiocytes). The spike protein subunits are of interest as targets of vaccines; the S2 subunit is highly conserved and may be an effective pan-coronavirus target. SARS-CoV studies in monkeys immunized with full-length spike protein showed successful protection against subsequent infection (Shang, 2020).
Camostat Mesylate
Camostat mesylate is an inhibitor of the protease TMPRSS2. This host protease is used by SARS-CoV-2 for spike protein priming, and inhibition with camostat mesylate has been shown to block SARS-CoV-2 infection in lung cell lines (Hoffmann, 2020).
Emodin
Emodin is an anthraquinone derivative extracted from Rheum tanguticum. It has been found to inhibit the SARS-CoV ORF3a protein and also block interactions between the SARS-CoV spike protein and the ACE2 receptor. It is of interest as a potential inhibitor of SARS-CoV-2 infection (Zhou, Y., 2020).
SARS-CoV-2 Membrane (M) Protein Inhibitors
The membrane (M) protein determines the shape of the SARS-CoV-2 viral envelope and organizes viral assembly through interaction with each additional structural protein (Chang, 2014).
Toremifene
Toremifene is a nonsteroidal selective estrogen receptor modulator (SERM) used in the treatment of metastatic breast cancer. It has also been found to block the fusion of viral and endosomal membranes by destabilizing the viral membrane glycoprotein and has been demonstrated to inhibit Ebola, MERS-CoV, and SARS-CoV viral replication in vitro using established cell lines. It is of interest as a potential inhibitor of SARS-CoV-2 infection (Zhou, Y., 2020).
SARS-CoV-2 Envelope (E) Protein Inhibitors
The envelope (E) protein is the smallest of the SARS-CoV-2 structural proteins, and it is incorporated into the virion envelope, though this represents only a small amount of total expressed envelope protein. A high proportion is also expressed inside the infected cell, where it is involved in viral assembly, budding, maturation, and propagation (Schoeman, 2019; Chang, 2020). The E protein sequence of SARS-CoV-2 has 90% amino acid overlap with other human coronavirus envelope proteins, which may allow for the development or repurposing of pan-coronavirus antiviral drugs that target this protein.
Belachinal, Macaflavanone E, and Vibsanol B are phytochemicals which have been shown in in silico models to potentially inhibit the activity of E protein (Gupta, M.G., 2020).
SARS-CoV-2 Papain-like Protease Inhibitors
There are two types of proteases expressed by SARS-CoV and SARS-CoV-2, the papain-like protease (PLpro, NSP3), and the CL-like protease (NSP5A & B, 3CLpro, Mpro). These proteases are responsible for cleaving the viral polyprotein and releasing nonstructural proteins (NSPs). These NSPs are vital for SARS-CoV-2 viral replication, maturation, and its overall life-cycle. In SARS-CoV, the papain-like protease (PLpro) inhibits type I interferon (IFN) by blocking IRF3 phosphorylation, which results in downstream inhibition of interferon induction and a reduction in the host’s innate immune response. This is thought to contribute to higher viral titer and leads to an increase in cell death and damage to infected and surrounding tissue (Matthews, 2014). The SARS-CoV-2 papain-like protease is thus of interest as a drug target to prevent viral replication and potentially reduce tissue damage (Báez-Santos, 2015).
Lopinavir
Lopinavir is the most powerful inhibitor of CoV protease and Saquinavir is the least powerful. As per current guidelines, Lopinavir + Ritonavir is the recommended protease inhibitor combination for the treatment of COVID-19.
Mercaptopurine (6MP)
Mercaptopurine has been found to inhibit both SARS-CoV and MERS-CoV papain-like protease (PLpro), a protein necessary for viral maturation. Mercaptopurine acts as an anti-inflammatory agent by inhibiting interferon stimulation via proteases (Zhou, Y., 2020; Chen, X., 2009; Cheng, K.W., 2015).
Darunavir
Darunavir is a protease inhibitor. When used in combination with cobicistat it has been found to inhibit the 3CL-protease (3CLpro), thereby blocking viral replication.
Oseltamivir (Tamiflu)
Oseltamivir is a neuraminidase inhibitor, a competitive inhibitor of influenza’s neuraminidase enzyme. The enzyme cleaves the sialic acid which is found on glycoproteins on the surface of human cells, which helps new virions to exit the cell. The use of oseltamivir in combination ASC09 or Ritonavir is currently under clinical study for treating coronavirus infections. (Harrison, C., 2020)
Nelfinavir
Nelfinavir is an HIV-1 protease inhibitor that has been demonstrated to also suppress SARS-CoV viral replication (Yamamoto, 2004). Nelfinavir is of interest in COVID-19 antiviral drug research, as it was found to inhibit the SARS-CoV-2 papain-like protease (plPro, mPro) when tested in Vero E6 cells (Xu, 2020). It has also been shown to inhibit inflammatory cytokines in vitro and may be effective in reducing the inflammatory response and severe innate immune activation resulting from COVID-19 and other viral diseases (Xu, 2020; Wallet, 2012).
SARS-CoV-2 NSP12 Polymerase Inhibitors
NSP12 RNA-dependent RNA polymerase (RdRP, RDR, RNA replicase) is an enzyme that catalyzes the replication of RNA from an RNA template. RdRP is a crucial viral enzyme in the life cycle of RNA viruses. In all positive-strand RNA viruses including SARS-CoV and SARS-CoV-2, RdRP constitutes the central catalytic subunit of the machinery involved in RNA synthesis and catalyzes the replication and transcription of the RNA genome (te Velthuis, 2010).
Acyclovir
Acyclovir is a synthetic nucleoside analog that serves as a polymerase inhibitor. Acyclovir fleximers have shown to inhibit HCov-NL63 and MERS-CoV (Creative Biolabs, 2020).
Baloxavir marboxil
Baloxavir marboxil is a Cap-dependent endonuclease inhibitor and is used in combination with favipiravir, a nucleoside analog RdRP polymerase inhibitor, for SARS-CoV-2 treatment (Harrison, C., 2020).
Favipiravir (T-705)
Favipiravir is an RNA polymerase inhibitor used for the treatment of Influenza A and B. Although it has not shown strong activity against SARS-CoV-2 (Wang, M., 2020), it is still in trial for use in combination with other therapies (Harrison, C., 2020).
Emtricitabine
Emtricitabine acts as a reverse transcriptase inbibitor. Although coronaviruses do not have reverse transcriptase, repurposing of these inhibitors is being explored in concert with other antivirals for treatment of COVID-19 (Harrison, C., 2020).
Tenofovir
Tenofovir acts as a guanosine analog reverse transcriptase inhibitor. In combination, Emtricitabine/Tenofovir inhibit viral RNA synthesis. These have been used in trials in combination with guanosine analogs and RNA synthesis inhibitors (Harrison, C., 2020).
Galidesivir (BCX4430, Immucillin-A)
Galidesivir tightly binds to RdRP and is used as a polymerase inhibitor (Elfiky, A.A., 2020).
Remdesivir
Remdesivir is a broad spectrum anti-viral developed against Ebola virus and is being tried as an anti-replicase.
Ribavirin
Ribavirin is an anti-polymerase drug which binds tightly to RdRP (Elfiky, A.A., 2020).
Sofosbuvir
Sofosbuvir is an anti-HCV drug which targets RdRP (Elfiky, A.A., 2020).
SARS-CoV-2 Open Reading Frame 3 (ORF3a) Inhibitors
The ORF3a protein expressed by SARS-CoV-2 has 72% sequence homology with SARS-CoV ORF3a. In SARS-CoV, the ORF3a protein activates NF-κB and the NLRP3 inflammasome by inducing TRAF3-dependent ubiquitination of p105 and ASC (Kam-Leung Siu, 2019). There is evidence that the SARS-CoV-2 virus is less effective in activating the NLRP3 inflammasome and in suppressing the antiviral response when compared to SARS-CoV. More studies are needed to fully understand how the SARS-CoV-2 ORF3a protein influences the immune and inflammatory response as it pertains to COVID-19 disease progression (Yuen, 2020; Zeng, 2004).
Tranilast
Tranilast inhibits activation of the NLRP3 inflammasome pathway, which is activated by ORF3a.
Helicase Inhibitors
The coronavirus helicase protein (nsp13) is conserved among coronavirus species, in contrast to structural proteins (e.g. spike) that show high sequence variability. Given the 70-80% sequence similarity of helicases among MERS, SARS, and SARS-CoV-2, and the effectiveness of drugs targeting protease and polymerase enzymes of MERS and SARS (Indinavir, Remdesivir), the helicase protein is therefore an attractive potential target for COVID-19 therapy.
Helicase enzymes are motor proteins that use energy derived from ATP hydrolysis (Briguglio I, 2010). Nsp13 helicase has been demonstrated to catalyze NTP-dependent unwinding of duplex oligonucleotides (RNA or DNA) into single strands, a step that is required for viral replication (Jia Z, 2019; Singleton MR, 2007).
Helicase inhibitors can have the following mechanisms of action:
- 1) Interfere with ATP binding to limit energy required from ATP hydrolysis
- 2) Inhibit NTPase activity by allosteric mechanisms
- 3) Inhibit nucleic acid binding to the helicase
- 4) Inhibit coupling of hydrolysis to unwinding
In silico studies on MERS and SARS-CoV have shown helicase domain 1A to have a direct effect on unwinding (Mirza MU, 2020). Small compounds that have the potential to inhibit ATP binding or direct NTPase activity include benzotriazole, imidazole, imidazodiazepine, phenothiazine, quinoline, anthracycline, triphenylmethane, tropolone, pyrrole, acridone, small peptide, and bananin derivatives (Briguglio I, 2010). Such compounds have been demonstrated to inhibit ATPase activity, leading to inhibition of viral replication in vitro, and may prove to be of benefit in vivo.
References
-
Báez-Santos YM et al. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015
-
Briguglio I, Piras S, Corona P, & Carta A. (2010). Inhibition of RNA Helicases of ssRNA + Virus Belonging to Flaviviridae, Coronaviridae and Picornaviridae Families. International Journal of Medicinal Chemistry 2011: 1–22. doi.org/10.1155/2011/213135
-
Chang, C. K., Hou, M. H., Chang, C. F., Hsiao, C. D., & Huang, T. H. (2014). The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Research, 103, 39–50.
-
Chen, X., Chou, C. Y., & Chang, G. G. (2009). Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antiviral Chemistry & Chemotherapy, 19(4), 151–156.
-
Cheng, K. W., Cheng, S. C., Chen, W. Y., Lin, M. H., Chuang, S. J., Cheng, I. H., Sun, C. Y., & Chou, C. Y. (2015). Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral Research, 115, 9–16.
-
Creative Bioloabs. (2020). Acyclovir Fleximer for the Treatment of SARS-CoV-2.
https://sars-cov-2.creative-biolabs.com/acyclovir-fleximer-for-the-treatment-of-sars-cov2.htm. -
Elfiky A. A. (2020). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 117592.
-
Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure & Dynamics, 1–11.
-
Harrison, C. (2020, Feb 20). Coronavirus puts drug repurposing on the fast track. Nature Biotechnology.
https://www.nature.com/articles/d41587-020-00003-1. -
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N. H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell, 181(2), 271–280.e8.
-
Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, Zheng L, Ming Z, Zhang L, Lou Z, & Rao Z. (2019). Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Research 47(12): 6538–6550. doi.org/10.1093/nar/gkz409
-
Kam-Leung Siu, Kit-San Yuen, Carlos Castaño-Rodriguez, Zi-Wei Ye, Man-Lung Yeung, Sin-Yee Fung, Shuofeng Yuan, Chi-Ping Chan, Kwok-Yung Yuen, Luis Enjuanes, and Dong-Yan Jin, The FASEB Journal 2019 33:8, 8865-8877
-
Matthews, K., Schäfer, A., Pham, A. et al. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol J 11, 209 (2014).
https://doi.org/10.1186/s12985-014-0209-9 -
Mirza MU, & Froeyen M. (2020). Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. Journal of Pharmaceutical Analysis 10(4): 320–328. doi.org/10.1016/j.jpha.2020.04.008
-
Schoeman, D., & Fielding, B. C. (2019). Coronavirus envelope protein: current knowledge. Virology Journal, 16(1), 69.
-
Shang, W., Yang, Y., Rao, Y., & Rao, X. (2020). The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines, 5, 18.
-
Singleton MR, Dillingham MS, & Wigley DB. (2007). Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annual Review of Biochemistry 76(1): 23–50. doi.org/10.1146/annurev.biochem.76.052305.115300
-
Siu, K. L., Yuen, K. S., Castaño-Rodriguez, C., Ye, Z. W., Yeung, M. L., Fung, S. Y., Yuan, S., Chan, C. P., Yuen, K. Y., Enjuanes, L., & Jin, D. Y. (2019). Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 33(8), 8865–8877.
-
te Velthuis, A. J., van den Worm, S. H., Sims, A. C., Baric, R. S., Snijder, E. J., & van Hemert, M. J. (2010). Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathogens, 6(11).
-
Wallet, M. A., Reist, C. M., Williams, J. C., Appelberg, S., Guiulfo, G. L., Gardner, B., Sleasman, J. W., & Goodenow, M. M. (2012). The HIV-1 protease inhibitor nelfinavir activates PP2 and inhibits MAPK signaling in macrophages: a pathway to reduce inflammation. Journal of Leukocyte Biology, 92(4), 795–805.
-
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell research, 30(3), 269–271.
-
Xu, Z., Yao, H., Shen, J., Wu, N., Xu, Y., Lu, X., zhu. w., & Li, L.-J. (2020). Nelfinavir Is Active Against SARS-CoV-2 in Vero E6 Cells. ChemRxiv. 12039888.v1 https://doi.org/10.26434/chemrxiv.12039888.v1.
-
Yamamoto, N., Yang, R., Yoshinaka, Y., Amari, S., Nakano, T., Cinatl, J., Rabenau, H., Doerr, H. W., Hunsmann, G., Otaka, A., Tamamura, H., Fujii, N., & Yamamoto, N. (2004). HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochemical and Biophysical Research Communications, 318(3), 719–725.
-
Yuen KS et al. Cell Biosci. 2020 Mar 16;10:40. doi: 10.1186/s13578-020-00404-4
-
Zeng, R., Yang, R. F., Shi, M. D., Jiang, M. R., Xie, Y. H., Ruan, H. Q., Jiang, X. S., Shi, L., Zhou, H., Zhang, L., Wu, X. D., Lin, Y., Ji, Y. Y., Xiong, L., Jin, Y., Dai, E. H., Wang, X. Y., Si, B. Y., Wang, J., Wang, H. X., … Wu, J. R. (2004). Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. Journal of Molecular Biology, 341(1), 271–279.
-
Zhou, Y., Hou, Y., Shen, J., Huang, Y., Martin, W., & Cheng, F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6, 14.
Coronavirus and SARS-CoV-2 Inhibitors
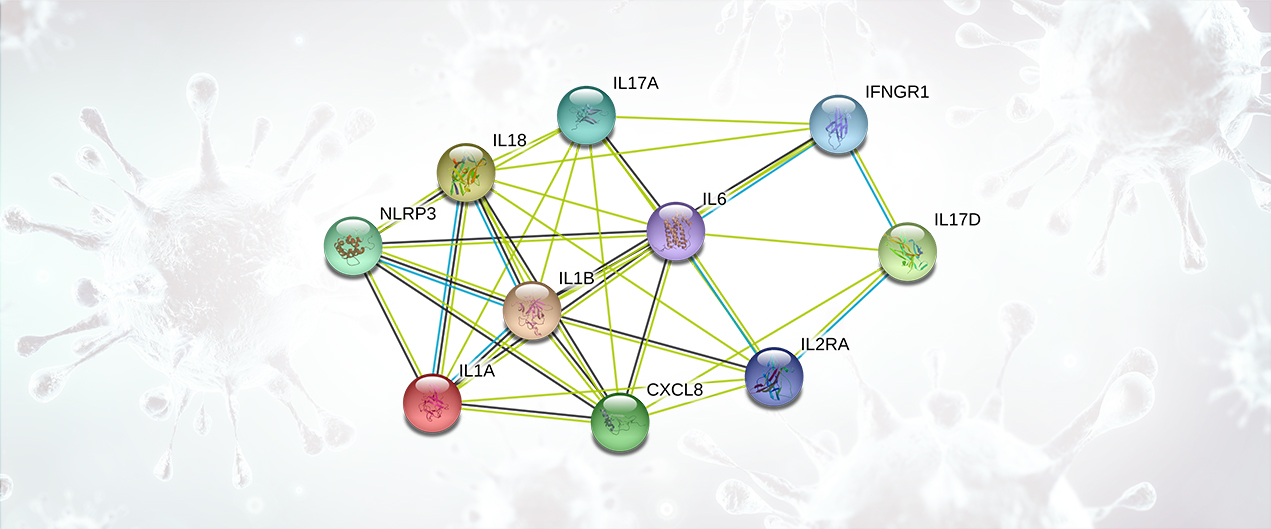
Cytokines are small proteins produced in the body to mediate cell signaling and regulation of the immune system. Cytokine Release Syndrome is characterized by an immune over-response, where the majority of the damage done to the host is due to an attack by the immune system rather than a direct cytotoxic effect by the pathogen. This systemic inflammatory response is mediated by the release (by immune cells) of inflammatory cytokines such as IFN-alpha, IFN-gamma, IL-1, IL-1 beta, IL-2, IL-2R, IL-6, IL-8, IL-10, IL-12, IL-17D, IL-18, IL-33, TNF-alpha, TGF-beta, and chemokines such as CCL2, CCL3, CCL5, CXCL8, CXCL9, and CXCL10. The overproduction of inflammatory cytokines leads to an attack on multiple organ systems resulting in ARDS, multi-organ failure, and hypotensive shock (Li, X., 2020; Huang, C., 2020). Both severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV) cause a severe and highly lethal respiratory disease in humans. These diseases are also characterized by a prominent pro-inflammatory response and mediated by release of cytokines (Chan, J.F., 2015).
Cytokine Release Syndrome Targets
Interleukin-1 Alpha (IL-1A)
IL-1A is a pro-inflammatory cytokine and an important mediator of local and systemic inflammation. Excessive IL-1A release during viral infections can cause lung and tissue inflammation, fever, and fibrosis. IL-1A suppression has been found to be effective in many inflammatory diseases, including rheumatoid arthritis.
Interleukin-1 Beta (IL-1B)
IL-1B is a potent pro-inflammatory cytokine. Initially discovered as the major endogenous pyrogen, it induces prostaglandin synthesis, neutrophil influx and activation, T-cell activation and cytokine production, B-cell activation and antibody production, and fibroblast proliferation and collagen production. It promotes differentiation of the TH17 subset of T-cells.
Interleukin-2 (IL-2)
Produced by T-cells in response to antigenic or mitogenic stimulation, this protein is required for T-cell proliferation and many other activities crucial to regulation of the immune response. It can stimulate B-cells, monocytes, lymphokine-activated killer cells, natural killer cells, and glioma cells.
Interleukin-2 Receptor (IL-2R)
The increased expression of IL-2R and IL-6 in serum is expected to predict increased severity of COVID-19 pneumonia and a poorer patient prognosis (Chen, L., 2020).
Interleukin-2 Receptor Gamma (IL-2RG)
IL-2RG is a common subunit for the receptors for a variety of interleukins. Probably in association with IL-15RA, it is involved in the stimulation of neutrophil phagocytosis by IL-15.
Interleukin-6 (IL-6)
IL-6 is a cytokine with a wide variety of biological functions. It is a potent inducer of the acute phase response, plays an essential role in the final differentiation of B-cells into immunoglobulin-secreting cells, and is involved in lymphocyte and monocyte differentiation. IL-6 acts on B-cells, T-cells, hepatocytes, hematopoietic progenitor cells, and cells of the central nervous system (CNS). It is also required for the generation of TH17 cells.
Increased expression of IL-2R, D-Dimer, and IL-6 in serum was associated with increased severity of 2019-nCoV pneumonia and poorer patient prognosis (Russell, B., 2020) (Chen, L., 2020).
Interleukin-8 (IL-8)
IL-8 is a chemotactic factor that attracts neutrophils, basophils, and T-cells, but not monocytes. It is also involved in neutrophil activation. IL-8 is released from several cell types in response to an inflammatory stimulus.
Interleukin-10 (IL-10)
IL-10 inhibits the synthesis of a number of cytokines, including IFN-gamma, IL-2, IL-3, TNF, and GM-CSF. IL-10 is produced by activated macrophages and by helper T-cells.
Interleukin-17D (IL-17D)
IL-17D induces expression of IL-6, CXCL8/IL-8, and CSF2/GM-CSF from endothelial cells.
Interleukin-17A (IL-17A)
IL-17A is part of the ligand for IL-17RA and IL-17RC. The heterodimer formed by IL-17A and IL-17F is the ligand for the heterodimeric complex formed by IL-17RA and IL-17RC. It is involved in inducing stromal cells to produce proinflammatory and hematopoietic cytokines.
Interleukin-18 (IL-18)
IL-18 augments natural killer cell activity in spleen cells and stimulates interferon gamma production in T-helper type I (TH1) cells. IL-18 belongs to the IL-1 family.
Interferon Gamma Receptor 1 (IFNGR1)
IFNGR1 associates with IFNGR2 to form the receptor for interferon gamma (IFNG). Ligand binding stimulates activation of the JAK/STAT signaling pathway.
Tyrosine-Protein Kinase JAK2
JAK2 is a non-receptor tyrosine kinase involved in various processes such as cell growth, development, differentiation, and histone modification. The JAK2 pathway mediates essential signaling events in both innate and adaptive immunity. In the cytoplasm, JAK2 plays a pivotal role in signal transduction via its association with type I receptors, such as growth hormone (GHR), prolactin (PRLR), leptin (LEPR), erythropoietin (EPOR), and thrombopoietin (TPOR); or with type II receptors, including IFN-alpha, IFN-beta, IFN-gamma, and multiple interleukins. JAK2 mediates angiotensin-2-induced ARHGEF1 phosphorylation, which may lead to the hypertension observed in some COVID-19 patients (Guilluy C, 2010).
Tumor Necrosis Factor-alpha (TNFa)
The TNF family of receptors and cytokines is very large and this family is frequently targeted by drugs. TNFa is a pro-inflammatory cytokine involved in autoimmune and immune-mediated disorders such as rheumatoid arthritis, ankylosing spondylitis, inflammatory bowel disease, psoriasis, hidradenitis suppurativa, and refractory asthma. TNFa inhibitors act by suppressing the physiologic response to TNFa. Inhibition of TNFa can be achieved with a monoclonal antibody, such as infliximab (Remicade), adalimumab (Humira), certolizumab pegol (Cimzia) and golimumab (Simponi), or with a receptor fusion protein, etanercept (Enbrel). Low-dose prednisolone and tacrolimus may have beneficial impacts on COVID-19 (Russell B., 2020).
NACHT, LRR and PYD domains-containing protein 3(NLRP3)
As the sensor component of the NLRP3 inflammasome, NLRP3 plays a crucial role in innate immunity and inflammation. The SARS-CoV open reading frame 3a (ORF3a) accessory protein activates the NLRP3 inflammasome by promoting TNF receptor-associated factor 3 (TRAF3)-mediated ubiquitination of apoptosis-associated speck-like protein, containing a caspase recruitment domain (ASC). Since the ORF3a domain in SARS-CoV-2 is approximately 73% similar to SARS-CoV, it is likely to be involved in a similar mechanism (Siu, K.L., 2019).
C-C motif chemokine 2 (MCP1/CCL2)
C-C motif chemokine 2 (MCP1/CCL2) is a chemotactic factor that attracts monocytes and basophils but not neutrophils or eosinophils. CCL2 augments monocyte anti-tumor activity, and has been implicated in the pathogenesis of diseases characterized by monocytic infiltrates like psoriasis, rheumatoid arthritis, and atherosclerosis.
Granulocyte Colony-Stimulating Factor (MIP1A/GCSF)
Granulocyte/macrophage colony-stimulating factors are cytokines that act during hematopoiesis by controlling the production, differentiation, and function of granulocytes, monocytes, and macrophages. GCSF induces granulocytes and belongs to the IL-6 superfamily.
References
-
Chan, J. F., Lau, S. K., To, K. K., Cheng, V. C., Woo, P. C., & Yuen, K. Y. (2015). Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clinical Microbiology Reviews, 28(2), 465–522.
-
Chen, L., Liu, H. G., Liu, W., Liu, J., Liu, K., Shang, J., Deng, Y., & Wei, S. (2020). Zhonghua jie he he hu xi za zhi = Zhonghua jiehe he huxi zazhi = Chinese Journal of Tuberculosis and Respiratory Diseases, 43(0), E005. Advance online publication.
-
Guilluy, C., Brégeon, J., Toumaniantz, G., Rolli-Derkinderen, M., Retailleau, K., Loufrani, L., Henrion, D., Scalbert, E., Bril, A., Torres, R. M., Offermanns, S., Pacaud, P., & Loirand, G. (2010). The Rho exchange factor Arhgef1 mediates the effects of angiotensin II on vascular tone and blood pressure. Nature Medicine, 16(2), 183–190
-
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., Xiao, Y., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England), 395(10223), 497–506.
-
Russell, B., Moss, C., George, G., Santaolalla, A., Cope, A., Papa, S., & Van Hemelrijck, M. (2020). Associations between immune-suppressive and stimulating drugs and novel COVID-19-a systematic review of current evidence. Ecancermedicalscience, 14, 1022.
-
Siu, K. L., Yuen, K. S., Castaño-Rodriguez, C., Ye, Z. W., Yeung, M. L., Fung, S. Y., Yuan, S., Chan, C. P., Yuen, K. Y., Enjuanes, L., & Jin, D. Y. (2019). Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB journal: Official Publication of the Federation of American Societies for Experimental Biology, 33(8), 8865–8877.
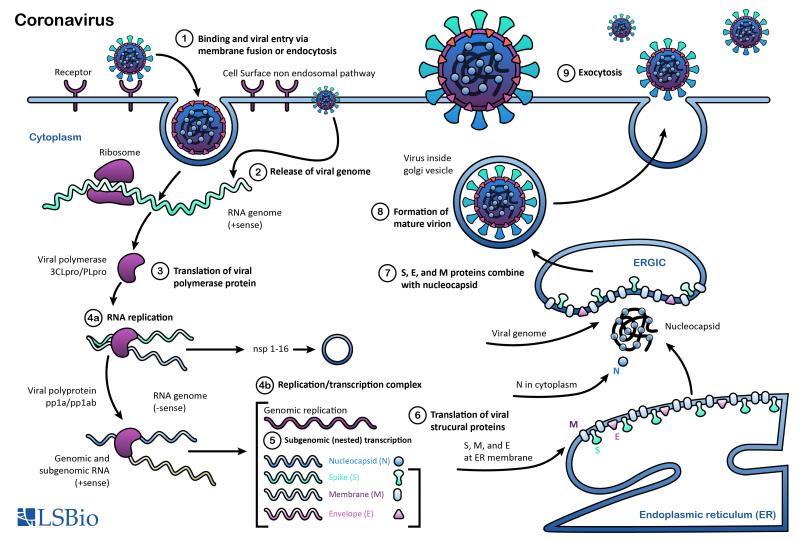
Coronaviruses (CoVs) are enveloped viruses with positive-sense, single-stranded RNA genomes that range in size from 26 to 32 kilobases, the largest among RNA viruses. They are pathogens known to infect humans and numerous other animal species. Based on genetic and antigenic criteria, CoVs have been organized into four groups: alpha, beta, gamma, and delta coronaviruses (Dhama, 2014; Coleman, 2014).
Common human coronaviruses include the 229E and NL63 alpha coronaviruses and the OC43 and HKU-1 beta coronaviruses. In infected humans, they are associated with a range of cold-like symptoms as well as severe respiratory tract infections (Fielding, 2011). Other more symptomatically severe human coronaviruses that have been transmitted from animals include MERS-CoV, a beta coronavirus that causes Middle East Respiratory Syndrome (MERS); SARS-CoV, a beta coronavirus that causes severe acute respiratory syndrome (SARS); and SARS-CoV-2, a beta coronavirus that causes coronavirus disease 2019 (COVID-19).
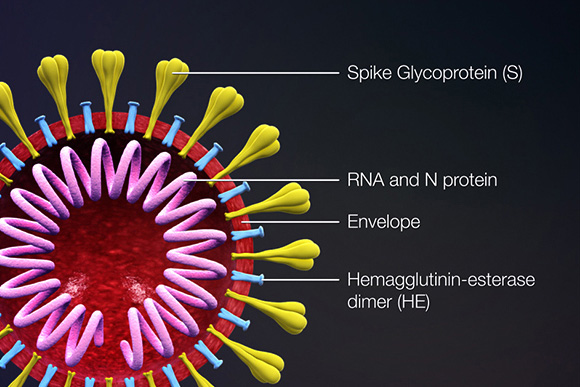
Proteins that contribute to the overall structure and function of all coronaviruses are the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, as well as the non-structural proteins cleaved from the orf1a/b polyprotein. Important non-structural proteins include the RNA-dependent RNA polymerase, protease, and endoRNAse (Fehr, 2016). The spike (S) protein is responsible for attachment to and fusion with the host cell. For both SARS-CoV and SARS-CoV-2, the host receptor is known to be Angiotensin converting enzyme 2 (ACE2) (Hoffman, 2020). The nucleocapsid (N) protein binds to the CoV genome and is responsible for packaging it into the ribonucleoprotein complex (capsid). The membrane (M) protein determines the shape of the viral envelope and organizes viral assembly through interaction with each additional structural protein. The envelope (E) protein is the smallest of the structural proteins and is incorporated into the virion envelope, but this represents only a small amount of total expressed envelope protein. A high proportion is also expressed inside the infected cell, where it is involved in viral assembly, budding, maturation and propagation (Schoeman, 2019; Chang, 2020).
The SARS-CoV-2 (COVID-19) coronavirus genome has 89% sequence similarity with the bat SARS-like CoVZXC21 coronavirus, and 82% similarity with human SARS-CoV coronavirus. The orf1a/b, S, E, M, and N proteins also share a high degree of phylogenetic similarity with bat, civet, and human SARS coronaviruses. Notable differences in sequence are found in the SARS-CoV-2 spike protein’s receptor binding domain, which has only 40% amino acid overlap with other related viruses. Furthermore, the SARS-CoV-2 orf3b protein represents a completely novel short protein (Rehman, 2020; Chan, 2020).
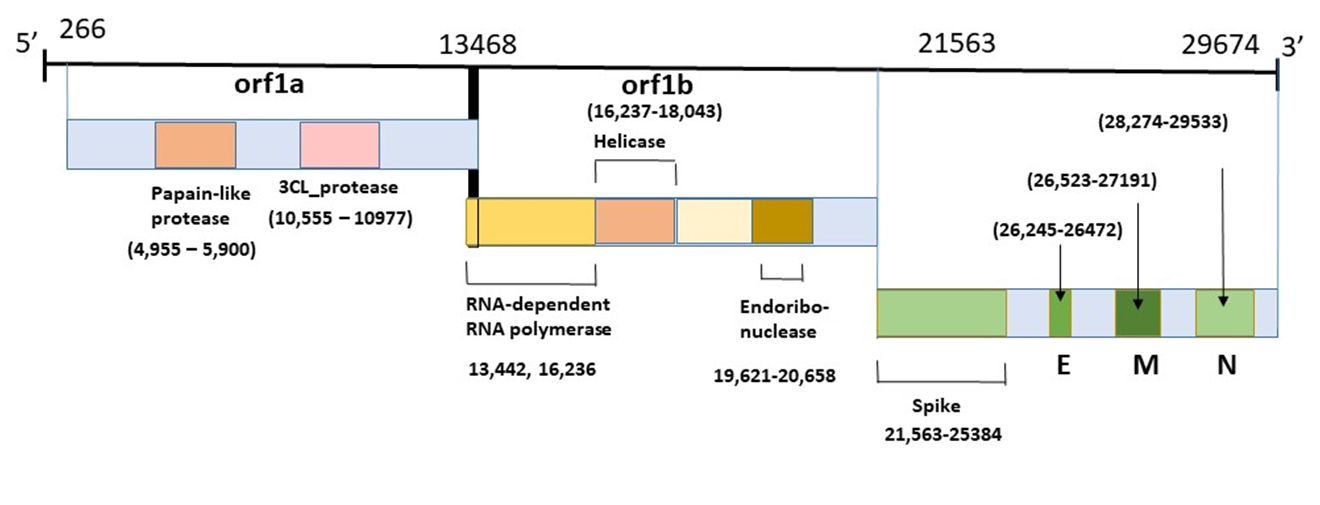
Coronavirus Genome
This diagram illustrates the genomic organization of the 29,674-nucleotides of the SARS-CoV-2 coronavirus. The regions that encode predicted ORFs 1a and 1b, which encode the nonstructural polyproteins (nsp1-16, helicase, 3CL-pro and PL-pro, RNA-dependent RNA polymerase), as well as the spike, envelope, membrane, and nucleocapsid structural proteins are indicated.
Coronavirus Spike Glycoprotein Phylogenetic Tree
This phylogenetic tree maps the evolutionary relationships of the SARS-CoV-2 Spike (S) glycoprotein with other coronaviruses. Analysis of the SARS-CoV-2 S protein indicated close proximity with the SARS-CoV and Bat RaTG13 coronaviruses, which are thought to be its closest relatives and potential origins. The next proximal relation is with the MERS coronavirus. Phylogenetic analysis is useful for coronavirus disease research and in the development of potential therapies, since drugs developed for closely related viruses (SARS-CoV, MERS) may be functionally repurposed as potential anti-SARS-CoV-2 agents.
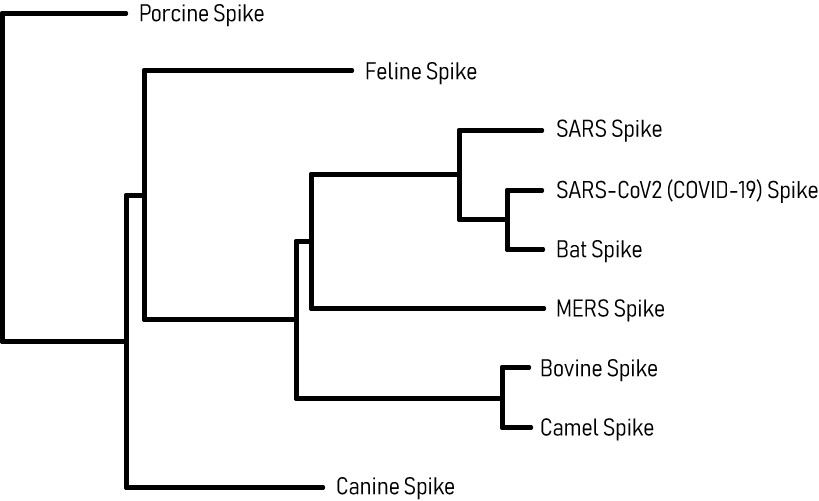
Coronavirus Family Spike Glycoprotein Homology Heatmap
The heatmap below displays the degree of sequence similarity of the SARS-CoV-2 Spike glycoprotein with that of other coronaviruses including SARS-CoV and MERS. It demonstrates that SARS-CoV-2 spike protein has high homology with the Bat RaTG13 and SARS-CoV Spike proteins, moderate similarity with bovine spike protein, and relatively low homology with canine coronavirus spike proteins.
Researchers worldwide are investigating SARS-CoV-2 virus, its mechanism of action, and how it causes COVID-19 disease. LSBio’s chief pathologist Dr. Glenna Burmer and bioinformaticians Mark Burmer and Vagmita Pabuwal have written a comprehensive review of SARS-CoV-2 and its involvement in the pathogenesis of COVID-19 disease. Topics include the viral life cycle, the phylogeny of coronaviruses, clinical features, pathogenesis and progression, inhibitors for treatment, and the efficacy of personal protective equipment. Read about SARS-CoV-2 and how it causes COVID-19 in the link below.