Inclusion of Super G in All Steps of Protein Arrays Improves S/N
Blocking reagents are used to reduce non-specific protein binding in immunoassays, significantly improving results in terms of Signal-to-Noise ratio. Typically the blocking reagent is added prior to addition of the primary detection antibody.
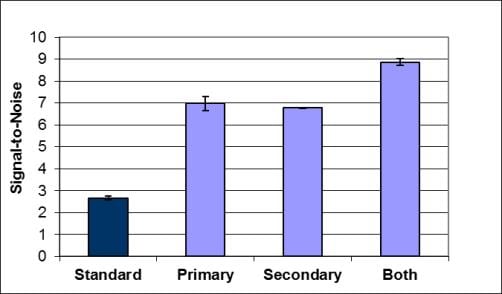 In this study we asked the question as to whether addition of the blocking reagent to other steps in a protein array assay has an impact on signal-to-noise results. We used Super G Blocking Buffer on our ONCYTE® nitrocellulose film slides for protein arrays using fluorescence detection and included the blocking buffer in various steps of the assay in addition to the ‘pre-assay’ step. Results show that addition of Super G Blocking Buffer to both antibody incubation steps and washing steps improved signal-to-noise
In this study we asked the question as to whether addition of the blocking reagent to other steps in a protein array assay has an impact on signal-to-noise results. We used Super G Blocking Buffer on our ONCYTE® nitrocellulose film slides for protein arrays using fluorescence detection and included the blocking buffer in various steps of the assay in addition to the ‘pre-assay’ step. Results show that addition of Super G Blocking Buffer to both antibody incubation steps and washing steps improved signal-to-noise
Grace Bio-Labs model protein array system is a serum cytokine sandwich immunoassay, using 1X Super G as pre-assay blocking agent, PBST + 0.5% BSA as a standard diluent for antibodies and PBST as the washing reagent. For comparison, Super G was included at 0.1x concentration in either the primary or secondary antibody incubations, or both incubations. Compared to the standard antibody dilution buffer, assays including 0.1x Super G in the incubation buffer resulted in an average increase in signal-to-noise for all cytokines tested of 2- to 3-fold for data collected at 635nm and 532nm, respectively (Figure 1).
In similar assays Grace Bo-Labs used only PBS/BSA as the antibody diluent, and 1x Super G was used in wash steps before primary antibody incubation, before the secondary antibody incubation, or before both incubations. Use of 1x Super G as a wash buffer (1 of 3 washes) resulted in an average increase in signal-to-noise for all cytokines tested of 2- to 3-fold for data collected at 635nm and 532nm, respectively (Figure 2).
The data support using Super G not only as a blocking reagent prior to immunoassays, but as a general component of the assay system when using nitrocellulose film slides. They recommend using Super G as a wash buffer between incubations (1x dilution) and as a component of incubation buffers (0.1x dilution) for optimal immunoassays on nitrocellulose film slides with fluorescence detection. Though these studies were conducted using only fluorescence detection, the implication is that inclusion of Super G Blocking Buffer could improve results with other detection methods such as colorimetric or chemiluminescence.
Figure 1. Super G as an Additive for Assay Incubation Diluent
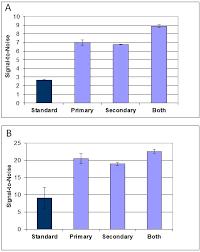
Figure 2. Super G as an Assay Wash Buffer
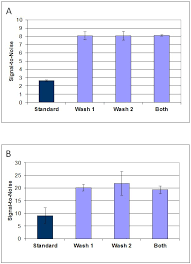
Figure Legends:
Figure 1. Signal-to-Noise obtained with a serum cytokine sandwich assay for IL1a, IL1b, IL6, TNFa, TNFb, IFNg comparing assays performed in PBST/0.5% BSA or the same buffer containing 0.1x Super G. Data are the mean ± standard deviations for n = 3 arrays. Results were obtained using detection at (A) 532nm and (B) 635nm using a Molecular Devices GenePix4000B Microarray Scanner. All arrays were blocked with 1x Super G prior to incubations. Standard = all incubations performed in PBST/0.5% BSA. Primary = only the detection antibody incubation was performed with PBST/0.5% BSA/0.1x Super G. Secondary = only the fluorescently labeled secondary antibody incubation was performed with PBST/0.5% BSA/0.1x Super G. Both = Both incubations were performed in the Super G containing buffer.
Figure 2. Signal-to-Noise obtained with a serum cytokine sandwich assay for IL1a, IL1b, IL6, TNFa, TNFb, IFNg comparing assays washed between incubations with Super G. Data are the mean ± standard deviations for n = 3 arrays. Results were obtained using detection at (A) 532nm and (B) 635nm using a Molecular Devices GenePix4000B Microarray Scanner. All arrays were blocked with 1x Super G prior to incubations and all incubations were performed in PBST/0.5% BSA. Standard = all washes were 3 x 5 minutes with PBST. Wash 1= only washes between the serum and the detection antibody incubations included 1x Super G. Wash 2 = only washes between the detection antibody and the secondary antibody incubations included 1x Super G. Both = All washes included Super G.