Nucleosome | Stratech
What is a nucleosome
Nucleosomes are integral components of all eukaryotic genomes with the purpose of organising and packaging DNA into small, condensed volumes to enable DNA to fit within a nucleus. Essentially, a nucleosome is a combination of a small section of DNA wrapped around a core of proteins called chromatin. This is an extremely efficient packaging system in which DNA is condensed into an 11 nm fibre that represents an approximate 6-fold level of compaction.
Over the last ten years or so, researchers have found more than one hundred unique histone post-translational modifications, a set of diverse chemicals chemical signatures that sit on the histone core.
Investigators are now increasingly coming to the belief that these modifications play a role in regulating chromatin architecture and could provide the basis for novel therapeutics. The vast majority of studies on chromatin either focus on acetylation or histone lysine methylation. These are accepted modes of research in the academic community and form the bedrock for work published in the academic literature.
This focus, however, is an artefact of the difficulty in precisely controlling the structure of the nucleosome. Researchers, however, are pioneering the development of a new approach called designer nucleosomes.
Designer nucleosomes are substrates that labs can use in a variety of assays, including the ever-popular ChIP (which we will cover below), nucleosome remodelling assays, enzymatic and binding assays and Western blot. While the original supply of designer nucleosomes was somewhat limited, that has now changed.
There are now advanced manufacturing processes that can create high quantities of these unique nucleosomes, providing more exotic methods for researchers to experiment with histone post-translational modifications (PTMs).
These advances are now opening exciting opportunities in the lab. These modifications have been shown to lead to lead to some either gene silencing or gene activation. The most common forms of modification include acetylation, methylation, or ubiquitination of lysine; methylation of arginine; and phosphorylation of serine. These modifications lead to epigenetic changes and has been investigated heavily in recent years.
Structure of a nucleosome
The structure of a nucleosome contains a segment of DNA wound around eight histone proteins and resembles beads-on-a-string (figure 1).
Histone proteins are a family of small, positively charged proteins termed H1, H2A, H2B, H3, and H4. DNA contains phosphate groups in its phosphate-sugar backbone leading to a negative charge, thus histones bind with DNA with high affinity. The nucleosome is the smallest structural component of chromatin.
The basic repeating structural (and functional) unit of chromatin is the nucleosome; the nucleosome contains a histone octamer protein consisting of 2 copies each of the core histones H2A, H2B, H3, and H4 with approximately 146 base pairs of DNA, wrapped in 1.67 left-handed super helical turns. Core particles are connected by stretches of linker DNA, which can be up to about 80 bp long as shown in figure 1.
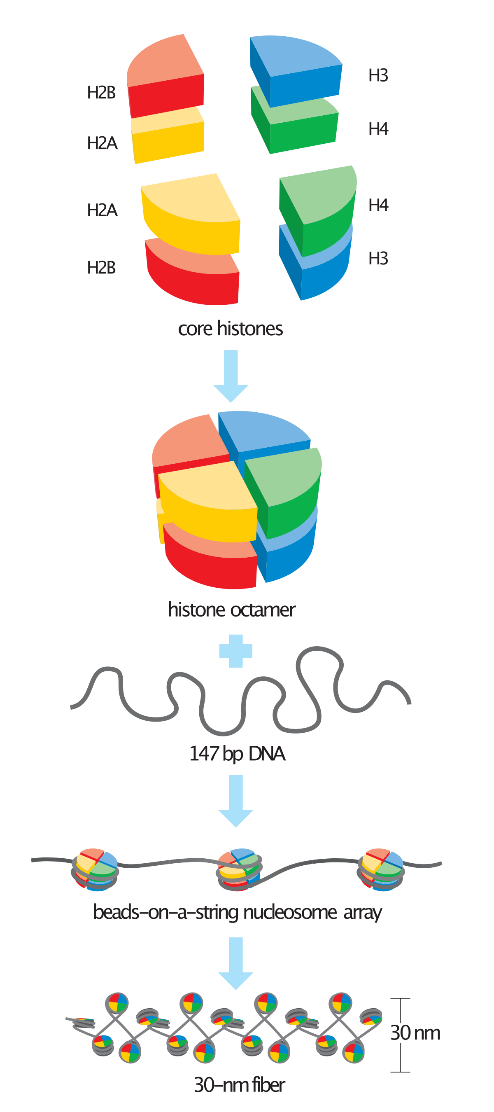
Figure 1: The histone octamer is formed from two subunits each of the four core histones. The four H3 and H4 subunits form a tightly packed tetramer that associates with two H2A/H2B dimers to form each octamer. About 147 bp of DNA is wrapped around the octamer to form a nucleosome. Nucleosomes can be arrayed in the loosely packed beads-on-a-string form of chromatin, but are generally more tightly packaged into the 30-nm fiber.
Fiber formation requires histone tails and additional proteins, neither of which is shown here.(Ref David O Morgan – The Cell Cycle. Principles of Control, 2007)
Histone Ubiquitination
Researchers have known since the 1970s that ubiquitination occurs on histone proteins. However, the importance of their role was poorly understood.
Today, however, there is an increasing demand for recombinant nucleosome modifications that involve histone proteins. The process of histone ubiquitination is essential for numerous intracellular pathways. Researchers have identified its relevance in responding to DNA damage and the role of chromatin-modifying enzymes in transcriptional regulation.
Histone ubiquitination also has ramifications for medical research. Investigators, for instance, now believe that alterations in histone ubiquitination are responsible for several chronic diseases, including Alzheimer’s and cancer. Thus, recombinant nucleosome substrates are essential tools for facilitating research into processes related to histone ubiquitination.
New Nucleosome Panels
Stratech suppliers are working on creating a broader range of recombinant nucleosomes for the many families of histone post-translational modifications. These families include those involved in citrullination, phosphorylation and ubiquitination. These suppliers are also investigating the use of nucleosomes for combinatorial post-translational modifications, opening new research possibilities.
Current technologies may not provide academic and commercial laboratories with the most rigorous approach. Research, for instance, suggests that dissecting an epigenetic mechanism against one to three nucleosomes is not a robust scientific methodology. Stratech, therefore, is working on a range of nucleosome panels that will allow researchers to track both on- and off-target interventions, whether investigating drugs, antibodies, or enzymes.
Stratech is adding new post-transcriptional modifications to its existing collection of nucleosomes for an ever-increasing range of experimental designs.
Enhanced Chromatin Profiling
Even though nearly two decades have passed since the profiling of the first human genome, scientific understanding of the genome is not complete. Researchers have deciphered the genome but are yet to comprehend the mechanisms entirely. Investigations have revealed that the function of DNA is mostly determined by its chromatin structure – how the spool of DNA coils and weaves inside the cell.
Thus, chromatin is vital in regulating how easily transcription factors and other proteins can access the DNA. Researchers now know that gene expression is not just related to the reading of the genome itself, but also the ability for proteins to access segments of the genome. Thus, enhanced chromatin profiling opens research into this area using assays. It is a novel method for investigating protein-DNA interactions and histone post-translational modifications that may be able to provide superior assay operation compared to traditional ChIP assays.
Antibodies
Epigenetic researchers currently struggled with poor specificity of histone post-transcriptional modification antibodies. Now, however, antibodies with extremely high specificity and sensitivity are available, offering opportunities for new lines of research and higher efficiency than ever before.
Nucleosome Panels
Nucleosome panels are tools that provide researchers with enhanced ability to study epigenetic information in biologically relevant nucleosome environments. The panels contain both single and combinatorially-modified nucleosomes. They come in both separate large panels as well as sets that focus on acylation and methylation targets.
Chromatin Remodelling Substrates
Understanding chromatin remodelling is essential for epigenetic research. Changes to chromatin within the cell is linked to a host of diseases, including cancer. To study chromatin effectively, however, scientists need highly defined nucleosomal substrates – both of which are difficult to adapt to existing assay technologies. Stratech offers a variety of chromatin remodelling substrates that are more effective than existing technologies.
Modified Designer Nucleosomes
Finding quality modified designer nucleosomes can be a challenge. Stratech offers a variety of semi-synthetic recombinant nucleosomes which contain histone post-transcriptional modifications. These nucleosomes are a better substrate for interactor proteins and the study of epigenetic enzymes.
Recombinant Nucleosomes
Recombinant nucleosomes are histone proteins from E.Coli, wrapped in synthetic Widom 601 DNA. These nucleosomes are a genuine biological substrate for naturally-occurring histone-modifying enzymes, making this type of nucleosome ideal for enzyme screening assays.
Oncogenic Nucleosomes
Cancer often leads to histone protein mutations. Until now, it has been difficult for researchers to study these in the physiological context. Oncogenic nucleosomes, however, change this dynamic. Oncogenic nucleosomes provide the most relevant nucleosomal substrates for cancer research.
Variant Nucleosomes
Variant nucleosomes are ideal for the study of processes in the cell involving the DNA replication, repair, and transcriptional regulation. They are a suitable physiological substrate for functional analysis of non-canonical changes to packets of DNA in eukaryotes.
Purified Nucleosomes
Purified nucleosomes are often described as the “ultimate experimental control.” Researchers use them as a positive control in Western blot, testing the binding activity of chromatin reader proteins, and as substrates for enzymatic reactions. These nucleosomes are free from any non-requisite DNA that might affect enzyme activities.
Nucleosome Subunits and Components
Sometimes researchers who are interested in studying nucleosomes do not require the entire biological component. Nucleosome subunits and components are parts of the nucleosome that have been isolated and prepared for experimentation. Nucleosome subunits include dimers, tetramers, Widom 601 sequence DNA and octamers.
Products available
Stratech Partner, Epicypher is Bringing Epigenetics to Life
Nucleosomes
Functionalized Recombinant Nucleosomes
H2A Ubiquitylated Mononucleosomes
Recombinant Proteins
EpiDyne® Chromatin Remodeling Enzymes
Epigenetics Reagents and Assays
CUTANA™ Ultra-sensitive genomic mapping assays for ChIC / CUT&RUN
EpiDyne™-FRET For Nucleosome Remodeling Assays
SNAP-ChIP™ Spike-in Controls for Quantitative ChIP
Epigenetics Compound Library
Inhibitor Screening library – DiscoveryProbe™ Epigenetics Compound Library