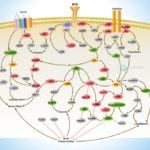
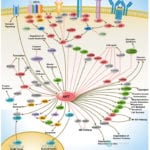
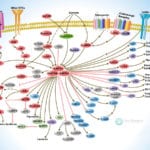
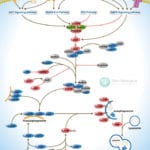
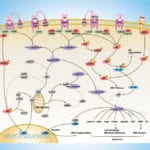
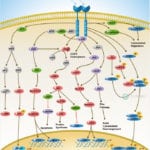
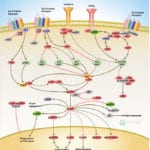
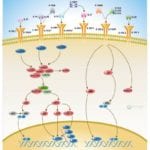
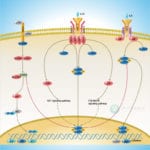
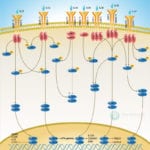
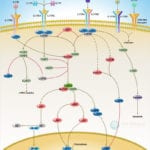
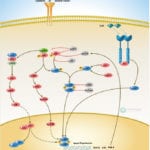
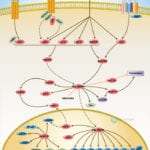
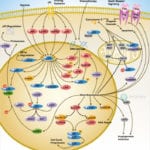
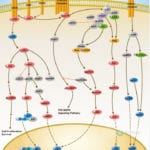
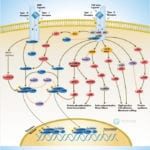
Browse or download our selection of Pathway Posters below:
Pathway Posters
Abeomics Pathways
- 14-3-3 Induced Apoptosis
- IL-18 Signaling
- Toll-Like Receptors
- DNA Methylation and Transcriptional Repression
- CMV and MAPK Pathways
- IL-6 receptor pathway
- NF-KappaB (p50-p65) Pathway
- Akt Signaling
- Apoptosis through Death Receptors
- Breast Cancer Regulation by Stathmin1
- Nanog in Mammalian ESC Pluripotency
- Oct4 in Mammalian ESC Pluripotency
- Caspase Cascade
- PD-1/PD-L1 Pathway in Cancer
- Cellular Apoptosis Pathway
- CD40 Signaling
- CD27 Pathway
- CTLA4 Signaling
- Cytokine Network
- TCR Signaling
- ZIKA Virus Infection And Host Response
- Allergic Response Pathway
- B-Cell Development
- Colorectal Cancer Metastasis
- Ebola Virus Pathogenesis
- EGF Pathway
- ErbB2-ErbB3 Heterodimers
- ERK Signaling
- Fas Signaling
- Human Embryonic Stem Cell Pluripotency
- IL-2 Gene Expression in Activated and Quiescent T-Cells
- IL-3 Signaling
- IL-17 Signaling
- miRNA in Cancer
- Mitochondrial Apoptosis
- Nucleotide Excision Repair Pathway
- Polyamine Regulation in Colon Cancer
- Rac1 Pathway
- Repair of Thymine Dimers
- TNF Signaling
- Tregs in Tumor Escape Checkpoints
- Activation of NF-KappaB by PKR
- Androgen Signaling
- APRIL Pathway
- ATM Pathway
- BRCA1 Pathway
- Calcium Mediated T-Cell Apoptosis
- c-Myc and Apoptosis
- CXCR4 Pathway
- p53 Mediated Apoptosis
- RANK Pathway
- TGF Beta Pathway
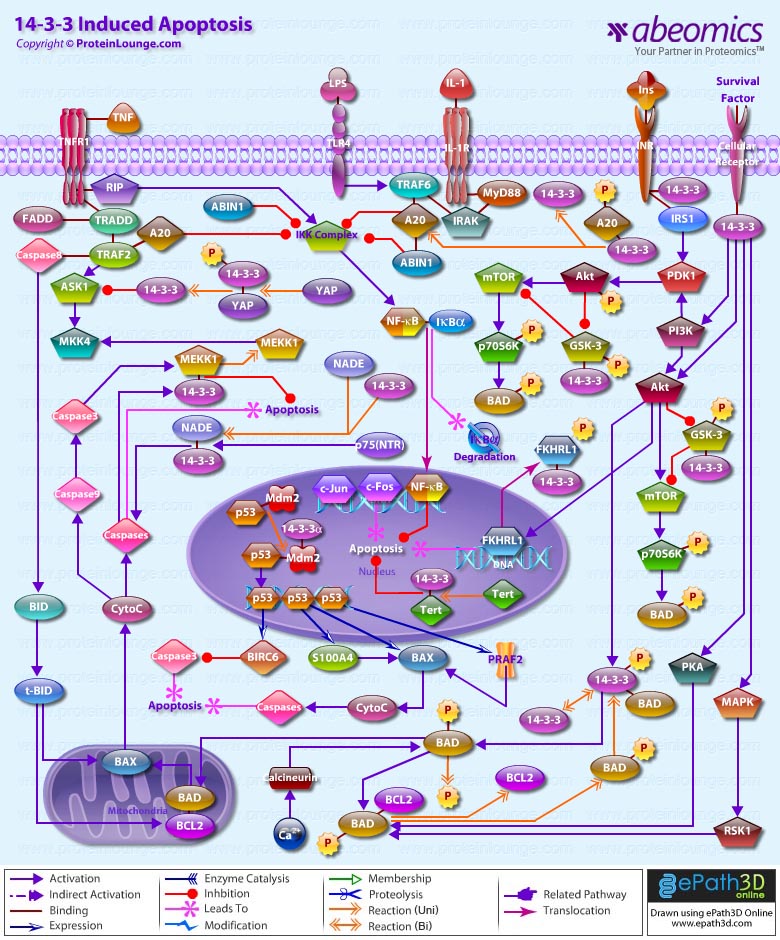
Apoptosis is a physiological process of cell death that plays a critical role in normal development as well as in the pathophysiology of a variety of diseases. The fundamental cellular mechanism behind apoptosis is due to a balance between anti-apoptotic and pro-apoptotic factors, which may be shifted by extracellular signals. 14-3-3 proteins play an important suppressing role in several apoptotic pathways in animals. These are a family of abundant, widely expressed 28-33-kDa acidic polypeptides that spontaneously self-assemble as dimmers. The 14-3-3 family of proteins mediates signal transduction by binding to phosphoserine-containing proteins. There are at least seven distinct genes for 14-3-3 in vertebrates, giving rise to nine isotypes (Alpha, Beta, Gamma, Delta, Epsilon, Eta, Sigma, Tau, and Zeta, with Alpha and Delta being phosphorylated forms of Beta and Zeta, respectively) (Ref.1).
Apoptosis can be initiated by ligand binding to members of the family of membrane bound TNF (Tumor Necrosis Factor) receptors, e.g., TNFR1 and CD95 (Fas). Activation of TNFR1 leads to recruitment of TRADD (TNF Receptor Associated Death Domain), which subsequently recruits the FADD (Fas-Associated Death Domain) and Caspase 8 (Ref.2). Caspase 8 directly activates Caspase 3 and BID (BH3-Interacting Domain death agonist), which activates BCL2 (B-Cell CLL/Lymphoma-2) in the mitochondria. Caspase 3 removes the N-terminal regulatory domain of MEKK1 (MAPK/ERK kinase kinase-1) from the catalytic domain, which ultimately results in apoptosis. TNF binding to TNFR1 also activates the recruitment of other TRADD, which via NIK (NF-KappaB-Inducing Kinase) ultimately activates the anti-apoptotic transcription factor NF-kappaB (Nuclear Factor-kappaB) complexed with inhibitory I-kappaB-Alpha (Inhibitor of Kappa Light Chain Gene Enhancer in B-Cells). The apoptosis-suppressing activity of NF-kappaB includes regulation of expression of the zinc finger protein A20 and anti-apoptotic genes (Ref.3). In addition, the binding of 14-3-3 to A20 mediates interaction with the c-Raf kinase, implicating a role for A20 in signal transduction and providing another 14-3-3-involving link between signal transduction and apoptosis (Ref.4). Intriguingly, 14-3-3 isoforms Eta and Zeta bind to the zinc finger domain of A20, which results in blocking other A20-binding inhibitor of NF-kappaB, ABIN1 (Nef-associated Factor-1) proteins from interacting with this domain. Moreover, A20 inhibits IKK (I-kappaB kinase) activation of NF-kappaB after apoptosis is prevented. With its nuclear localization signal, exposed NF-kappaB translocates to the nucleus, where it binds to kappaB sites in the promoter region of NF-kappaB -responsive genes. In addition, IL-1 (Interleukin-1) and LPS (Lipopolysaccharides) up-regulate A20 in endothelial cells, via a pathway similar to the TNF pathway, converging at the IKK complex (Ref.3). This involves the binding of IL-1/LPS to IL-1R/TLR4 (Toll-Like Receptor-4), which triggers a common intracellular signaling cascade involving the adapter proteins MyD88, TRAF6 (TNF Receptor-Associated Factor-6) and IRAK (IL-1R-Associated Kinase).
The kinase PI3K (Phosphatidylinositol-3 Kinase) and Akt1 exerts its anti-apoptotic role at several points in the apoptotic machinery. Besides phosphorylating BAD (BCL2 Associated Death Promoter) and FKHRL1 (Forkhead), Akt1 also inactivates Caspase 9 by phosphorylation on the Ser196 residue. ASK1 (Apoptosis Signal-regulating Kinase-1) associates with TRAF2 (TNF-R-Associating Factor-2), and mediates activation of c-Jun and c-Fos via the MKK4 (MAP Kinase Kinase-4)-JNK (Jun N-terminal kinase) pathway, resulting in TNF expression. Phosphorylation and 14-3-3 binding to ASK1 result in inhibition of the pathway (Ref.5). ASK1 appears to be a general mediator of cell death because it is responsive to a variety of additional death signals, including oxidative stress and treatment with the chemotherapeutic drugs cisplatin and paclitaxel. BAD inhibits the anti-apoptotic functions of BCL2 and BCL-XL but as a consequence of consecutive phosphorylation by Akt1 and PKA (Protein Kinase-A), BAD is released from BCL2 or BCL-XL and loses its pro-apoptotic effect. Conversely, pro-apoptotic phosphatases such as Calcineurin and PP2A (Protein Phosphatase 2A) dephosphorylate BAD, causing release from 14-3-3 proteins (Ref.6). Once phosphorylated, BAD can be complexed by 14-3-3 proteins in the cytoplasm, preventing the association of BAD with the mitochondrially localized BCL-XL and BCL2 and therefore inhibiting apoptosis. Similarly, phosphorylated FKHRL1 associates with 14-3-3 proteins and is therefore retained in the cytoplasm. On deprivation from survival signals, however, dephosphorylation of FKHRL1 leads to its dissociation from 14-3-3 and translocation to the nucleus, resulting in the subsequent activation of apoptotic genes like FasL (Ref.7).
14-3-3 proteins are abundantly expressed in the brain and have been detected in the cerebrospinal fluid of patients with different neurological disorders. By their interaction with more than 100 binding partners, 14-3-3 proteins modulate the action of proteins that are involved in regulation of cell cycle arrest in response to DNA damage, cell cycle timing, intracellular trafficking, regulation of ion channels, and intracellular signaling in response to stress, mating pheromone in yeast, photoreceptor development and learning in Drosophila, cellular response to stress and survival factors in mammals, and the Ras/Raf signaling pathway in various organisms (Ref.8).
References
1.14-3-3 proteins–an update.
Mhawech P.
Cell Res. 2005 Apr;15(4):228-36.
2.Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins.
Halbach T, Scheer N, Werr W.
Nucleic Acids Res. 2000 Sep 15; 28(18): 3542-50.
3.A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis.
Beyaert R, Heyninck K, Van Huffel S.
Biochem Pharmacol. 2000 Oct 15; 60(8): 1143-51. Review.
4.14-3-3 proteins: a number of functions for a numbered protein.
Bridges D, Moorhead GB.
Sci STKE. 2005 Aug 9;2005(296):re10.
5.Role of 14-3-3 proteins in eukaryotic signaling and development.
Darling DL, Yingling J, Wynshaw-Boris A.
Curr Top Dev Biol. 2005;68:281-315.
6.Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin- 3-dependent lymphoid cells by a mechanism requiring 14-3-3 dissociation.
Chiang CW, Harris G, Ellig C, Masters SC, Subramanian R, Shenolikar S, Wadzinski BE, Yang E.
Blood. 2001 Mar 1; 97(5): 1289-97.
7.Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis.
Basu S, Totty NF, Irwin MS, Sudol M, Downward J.
Mol Cell. 2003 Jan; 11(1): 11-23.
8.JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3.
Sunayama J, Tsuruta F, Masuyama N, Gotoh Y.
J Cell Biol. 2005 Jul 18;170(2):295-304. Epub 2005 Jul 11.
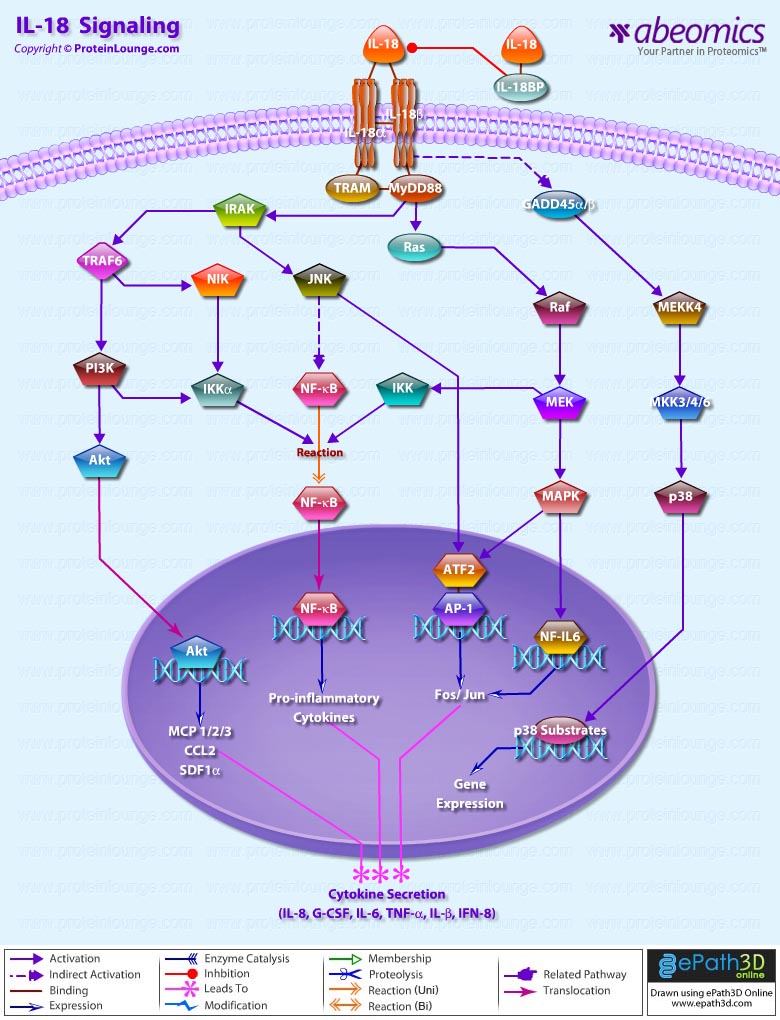
IL-18 is a proinflammatory cytokine that belongs to the IL-1 family of ligands. Macrophages and dendritic cells are the primary sources for active IL-18, but the IL-18 precursor is constitutively expressed in epithelial cells throughout the body. Similar to IL-1beta, IL-18 is synthesized as an inactive precursor requiring processing by caspase-1 into an active cytokine but unlike IL-1beta, the IL-18 precursor is constitutively present in nearly all cells in healthy humans and animals. The IL-18 receptors, although distinct from IL-1 receptors, also belong to the IL-1 receptor family. The IL-18 receptor (IL-18R) complex consists of two receptor chains; a ligand-binding chain termed the IL-18Ralpha chain and a co-receptor termed IL-18Rbeta chain; both chains are required for signaling (Ref.1 and 2).
The activity of IL-18 is balanced by the presence of a high affinity, naturally occurring IL-18 binding protein (IL-18BP). In humans, increased disease severity can be associated with an imbalance of IL-18 to IL-18BP such that the levels of free IL-18 are elevated in the circulation (Ref.3). IL-18 activates PI3K/Akt and MEK/ERK1/2 signaling pathways, contributing to the production of the chemokine MCP-1, which plays an important role in the recruitment of leukocytes at the inflammatory site and subsequently the development of adaptive immunity (Ref.4). By binding to IL-18RA, IL-18 upregulates IL-1R-associated kinase (IRAK) and TRAF-6 thus, results in nuclear translocation of nuclear factor kappa-B (NF-kB) and also activates p38 mitogen activated protein kinase (p38 MAPK) signaling (Ref.5). IL-18 has been implicated in several autoimmune diseases, myocardial function, emphysema, metabolic syndromes, psoriasis, inflammatory bowel disease, hemophagocytic syndromes, macrophage activation syndrome, sepsis, and acute kidney injury, although in some models of disease, IL-18 is protective (Ref.3).
References
1. Interleukin 1 and interleukin 18 as mediators of inflammation and the aging process.
Dinarello CA. Am J Clin
Nutr. 2006 Feb;83(2):447S-455S. Review.
2. Differences in signaling pathways by IL-1beta and IL-18.
Lee JK, Kim SH, Lewis EC, Azam T, Reznikov LL, Dinarello CA.
Proc Natl Acad Sci U S A. 2004 Jun 8;101(23):8815-20. Epub 2004 May 25.
3.Interleukin-18 and IL-18 binding protein.
Dinarello CA, Novick D, Kim S, Kaplanski G.
Front Immunol. 2013 Oct 8;4:289. doi: 10.3389/fimmu.2013.00289. Review.
4.IL-18 induces monocyte chemotactic protein-1 production in macrophages through the phosphatidylinositol 3-kinase/Akt and MEK/ERK1/2 pathways.
Yoo JK, Kwon H, Khil LY, Zhang L, Jun HS, Yoon JW.
J Immunol. 2005 Dec 15;175(12):8280-6.
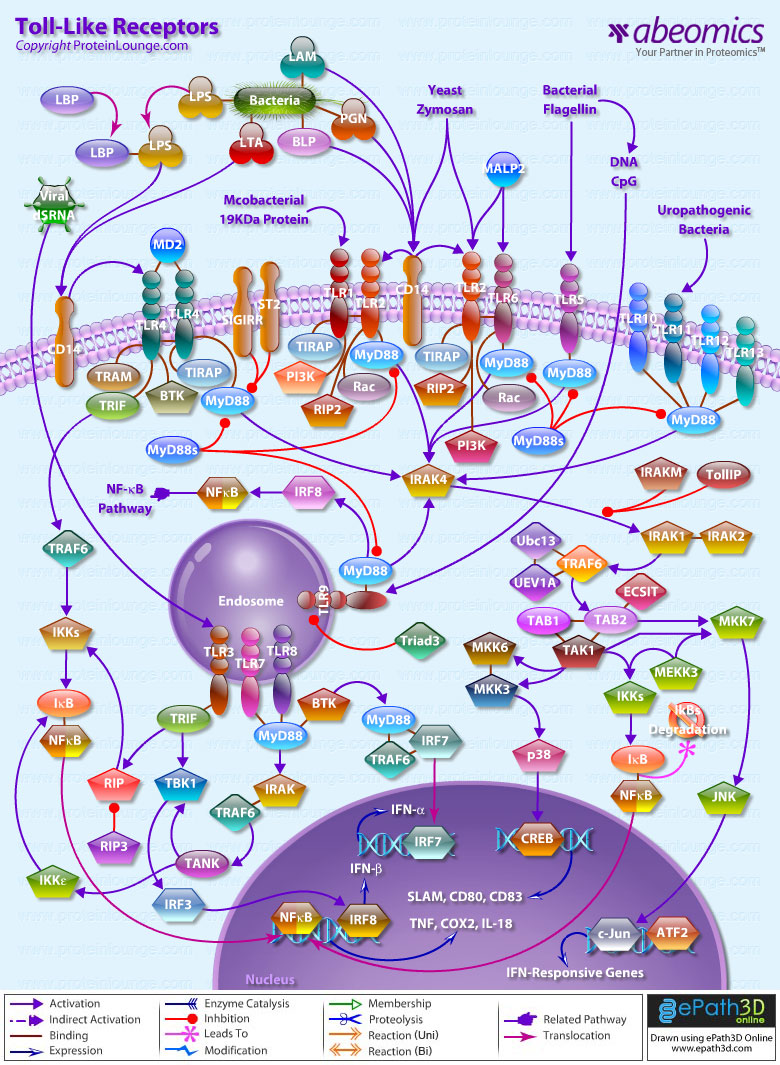
TLRs (Toll-like receptors) are transmembrane proteins expressed by cells of the innate immune system, which recognize invading microbes and activate signaling pathways that launch immune and inflammatory responses to destroy the invaders. Toll receptors were first identified in Drosophila. In mammals, the TLR family includes eleven proteins (TLR1-TLR11). Recently, two new members, and have been discovered in mouse, but not much information is known about them. Mammalian TLRs consist of an extracellular portion containing Leucine-rich repeats, a Transmembrane region and a Cytoplasmic tail, called the TIR (Toll-IL-1R (Interleukin-1-Receptor)) homology domain. Different TLRs serve as receptors for diverse ligands, including Bacterial cell wall components, Viral double-stranded RNA and small-molecule antiviral or immunomodulatory compounds (Ref.1). Activation of TLRs occurs after binding of the cognate ligand. Upon activation, TLRs activates two major signaling pathways. The core pathway activated by most TLRs leads to activation of the transcription factor (Nuclear Factor-KappaB) and the MAPKs (Mitogen-Activated Protein Kinases) p38 and JNK (c-Jun Kinase). The second pathway is activated by (Toll-Like Receptor-3) and (Toll-Like Receptor-4) and leads to activation of both and another transcription factor (Interferon Regulatory Factor-3), allowing for an additional set of genes to be induced, including antiviral genes such as Ifn- Beta (Interferon-Beta). In this way, TLRs can tailor the innate response to pathogens. TLRs that recognize nucleic acids signal from endosomes, whereas cell-surface TLRs sense lipids and proteins. Plasma membrane localized TLRs include TLR1, TLR2, TLR4, TLR5, and TLR10. Newly discovered TLR11, and are also believed to be Plasma membrane localized, whereas endosomal TLRs include TLR3, TLR7, and (Ref.2).
Mammalian is the signal-transducing receptor activated by the bacterial LPS (Lipopolysaccharide) and(Lipotechoic Acid). Both LPS andfirst bind to the (Cluster of Differentiation-14) receptor, which then transfer them to TLR4. homodimerizes and forms a complex with the protein MD2, and this complex is then transported to the cell surface. Thecan bind directly to CD14, but LPS must be delivered to by (LPS-Binding Protein). Cells need both and in order to recognize LPS. activation engages a set of (Myeloid Differentiation Primary-Response Protein-88) adaptor family members, including MyD88, (TIR Domain-containing Adapter Protein), TRIF, and TRAM. (Bruton Agammaglobulinemia Tyrosine Kinase) also participate in TLR signaling, although their precise role has yet to be defined (Ref.3). The adaptor protein link to the (Interleukin-1 Receptor-associated Kinase-1) and Serine/Threonine Kinases, leading to MyD88-dependent pathway. Upon activation of with LPS, recruits IRAK4, thereby allowing the association of IRAK1. then induces the phosphorylation of IRAK1. (Tumour Necrosis Factor-Receptor-Associated Factor-6) is also recruited to the receptor complex, by associating with phosphorylated IRAK1. Phosphorylated and then dissociate from the receptor and form a complex with (TGF-Beta-Activated Kinase-1), (TAK1-Binding Protein-1) and (TAK1-Binding Protein-2), which induces the phosphorylation of and TAK1. TRAF6, TAK1, and associate with the Ubiquitin ligases (Ubiquitin-Conjugating Enzyme-13) and (Ubiquitin-conjugating Enzyme E2-Variant-1). This leads to the ubiquitylation of TRAF6, which induces the activation of by interacting with (Evolutionarily Conserved Signaling Intermediate in Toll Pathways). TAK1, in turn, phosphorylates both p38 Kinases and JNKs by activating (Mitogen-Activated Protein Kinase Kinase-3), and MKK7. p38 and JNKs then enter the nucleus and induce the expression of their target genes. also phosphorylates the IKK complex (Inhibitor of Kappa Light Polypeptide Gene Enhancer in B-Cells Kinase), which consists of (Inhibitor of Kappa Light Polypeptide Gene Enhancer in B-Cells Kinase of Alpha), (Inhibitor of Kappa Light Polypeptide Gene Enhancer in B-Cells Kinase of Beta) and (Inhibitor of Kappa Light Polypeptide Gene Enhancer in B-Cells Kinase of Gamma). The IKK complex then phosphorylates I-KappaB, which leads to its ubiquitylation and subsequent degradation. This allows to translocate to the nucleus and induce the expression of its target genes. and link to pathways that lead to (TANK Binding Kinase-1) and activation (i.e., the MyD88-independent pathway) (Ref.1 & 4).
TLR2 is activated by bacterial LAM (Lipoarabinomannan), BLP (Bacterial Lipoprotein), and PGN (Peptidoglycans). LAM and PGN act on through the receptor, similar to the process followed by the with a similar downstream affect. BLP mediates both apoptosis and activation through TLR2. is also responsible for the recognition of the Yeast cell-wall particle Zymosan. Zymosan acts through the receptor to influence TLR2. signals the production of (Tumour Necrosis Factor), through pathway, from the phagocytized vesicle. associates with and recognizes diacylated (Mycoplasmal macrophage-Activating Lipopeptide-2 kD) along with TLR2. Like TLR4, they also signal through and TIRAP. (Phosphatidylinositde-3 Kinase), (Receptor-Interacting Protein-2) and Rac (Ras-Related Botulinum Toxin Substrate) are also involved in TLR6-TLR2 mediated signaling. also associates with and recognizes the native mycobacterial 19-kDa lipoprotein along with TLR2. TLR1-TLR2 also signals through MyD88, TIRAP, PI3K, and Rac. and may participate in the activation of Macrophages by Gram-positive bacteria. is a signaling mediator of bacterial Flagellin, thus activating and may play a role in resistance to Salmonella infection (Ref.5). Human is an orphan member of the TLR family. Genomic studies indicate that is in a locus that also contains and TLR6, two receptors known to function as coreceptors for TLR2. not only homodimerize but also heterodimerize with TLRs 1 and 2. It has been found to activate gene transcription through (Ref.6). is responsible for the recognition of CpG islands of bacterial DNA. The extracellular CpG fragment may activate TLR9, thus inducing the endocytosis of the DNA along with TLR9, or perhaps the bacteria is phagocytized and TLR9, which has separately formed on the phagosome, is activated by the CpG islands; which ever the exact method, activates the pathway from the endocytized vesicle. Recently (Interferon Regulatory Factor-8) has been shown to be activated by through (Ref.5).
Besides TLR9, three other TLRs found in endosome are TLR3, and TLR8. activates immune cells in response to double-stranded Viral RNA. The stimulation of the triggers activation that activates through TBK1, independent of MyD88, which lead to the secretion of Ifn-Beta. also activates RIP1 (Receptor-Interacting Protein-1) and TRAF6, which may further activate pathway. Small antiviral compounds activate immune cells via the TLR7 dependent signaling pathway. binds with and activates IRAF and TRAF6. then activates (also known as I-TRAF). interacts with and to activate IRF3. or may also activate through activation of MyD88, and TRAF6, thus inducing antiviral responses by producing (Interferon-Alpha). Recently, Mouse has been identified as a participant in defense against Uropathogenic bacteria. The ligands for Mouse and are currently unknown. Three of them are believed to signal through (Ref.7 & 8).
TLR signaling is also negatively regulated by various proteins. The cell-surface receptors (also known as T1) and (Single ImmunoGlobulin IL-IR-Related molecule (TIR-8)) function as inhibitory receptors, sequestering proteins from signaling complexes and preventing TLR2, and signaling. (Interleukin-1 Receptor-associated Kinase-M), (Toll-Interacting Protein) and a splice variant of MyD88, known as MyD88s, probably interfere with the recruitment and activation of and IRAK1. Recently, Triad3A, a RING-finger E3 ligase, has been shown to promote ubiquitylation of and TLR9, targeting these TLRs for degradation and thereby negatively regulating the intensity and duration of TLR signaling. The balance between activation and inhibition is the key determinant of signal strength of TLR pathways (Ref.9 & 10). In addition to the innate immune response, evidence implicates the involvement of the TLR family in a spectrum of systemic disorders following bacterial infections including Sepsis, Cardiac Ischemia, Peridontitis, and Cerebral palsy. The TLRs that control the onset of an acute inflammatory response are critical antecedents for the development of adaptive acquired immunity. Genetic and developmental variation in the expression of microbial pattern recognition receptors may affect the individual’s predisposition to infections in childhood and may contribute to susceptibility to severe neonatal inflammatory diseases, allergies, and autoimmune diseases (Ref.11).
References:
1.Inferences, questions and possibilities in Toll-like receptor signalling. Beutler B. Nature. 2004 Jul 8;430(6996):257-63.
2.Ligand-dependent Toll-like receptor 4 (TLR4)-oligomerization is directly linked with TLR4-signaling. Nishiya T, DeFranco AL. J Endotoxin Res. 2004;10(4):257-60.
3.The Toll-like receptor 2 is Recruited to Macrophage Phagosomes and Discriminates between Pathogens. Saitoh S, Akashi S, Yamada T, Tanimura N, Matsumoto F, Fukase K, Kusumoto S, Kosugi A, Miyake K. Nature: Dec.16, 1999: Vol.402, pp.39-43.
4.[TIR domain–containing adaptors regulate TLR-mediated signaling pathways] Yamamoto M, Akira S. Nippon Rinsho. 2004 Dec;62(12):2197-203. Review.
5.Toll-like receptors as sensors of pathogens. Hallman M, Ramet M, Ezekowitz RA.Pediatr Res 2001 Sep; 50(3): 315-21.
6.Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88.
Hasan U, Chaffois C, Gaillard C, Saulnier V, Merck E, Tancredi S, Guiet C, Briere F, Vlach J, Lebecque S, Trinchieri G, Bates EE. J Immunol. 2005 Mar 1;174(5):2942-50.
7.Linking Toll-like receptors to IFN-alpha/beta expression. Barton GM, Medzhitov R. Nat Immunol. 2003 May;4(5):432-3.
8.Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-{alpha} induction.
Uematsu S, Sato S, Yamamoto M, Hirotani T, Kato H, Takeshita F, Matsuda M, Coban C, Ishii KJ, Kawai T, Takeuchi O, Akira S. J Exp Med. 2005 Mar 21;201(6):915-23. Epub 2005 Mar 14.
9.RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. Nat Immunol. 2004 May;5(5):503-7. Epub 2004 Apr 4.
10.ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance.Brint EK, Xu D, Liu H, Dunne A, McKenzie AN, O’Neill LA, Liew FY. Nat Immunol. 2004 Apr;5(4):373-9. Epub 2004 Mar 7.
11.Toll-like receptors in the pathogenesis of human disease. Cook DN, Pisetsky DS, Schwartz DA. Nat Immunol. 2004 Oct;5(10):975-9. Review.
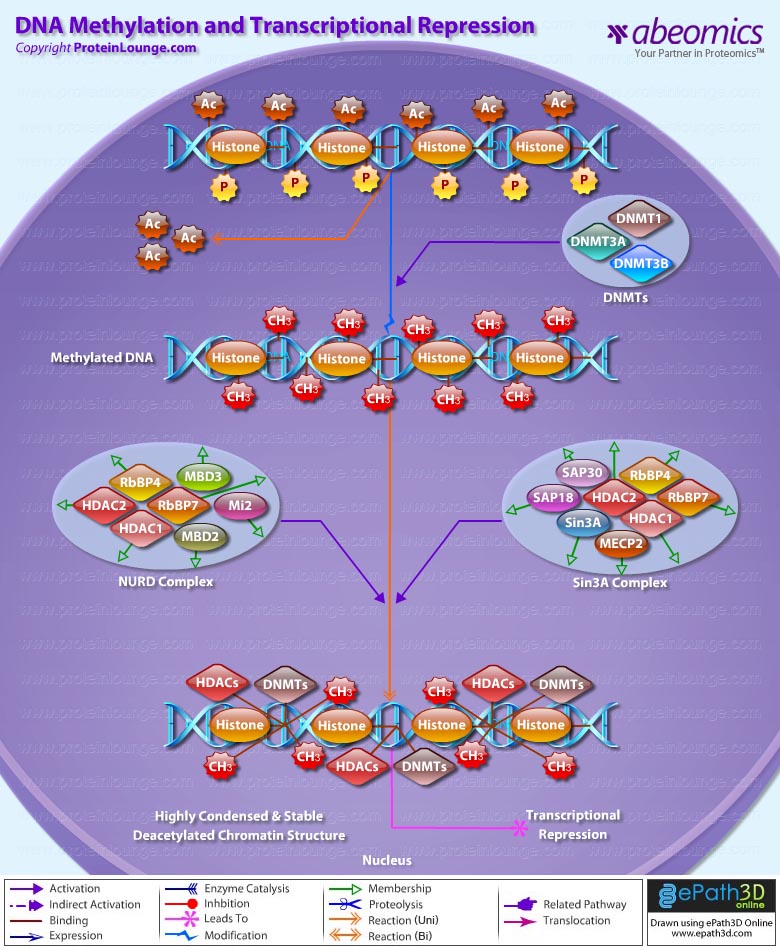
Transcriptional repression is an essential mechanism in the precise control of gene expression. Transcriptional repressor proteins associate with their target genes either directly through a DNA-binding domain or indirectly by interacting with other DNA-bound proteins. To inhibit transcription in a selective manner, a repressor protein can (1) mask a transcriptional activation domain, (2) block interaction of an activator with other components of the transcription machinery, or (3) displace an activator from the DNA. Furthermore, DNA response elements can exert allosteric effects on transcriptional regulators, such that regulators may activate transcription in the context of one gene, yet repress transcription in another (Ref.1).
The most direct mechanism by which DNA methylation can interfere with transcription is to prevent the binding of basal transcriptional machinery or ubiquitous TF (Transcription Factors) that require contact with cytosine (C) in the major groove of the double helix. Transcriptionally active chromatin is predominantly unmethylated and has high levels of acetylated histone tails. Most mammalian TFs have GC-rich binding sites and many have CpGs in their DNA recognition elements. Binding by several of these factors is impeded or abolished by methylation of CpG (Ref.2). Methylation at CpG dinucleotides is carried out by one of the three known human DNA methyltransferases (DNMT1, DNMT3a and DNMT3b), resulting in DNA with high levels of CpG methylation, but still containing predominantly acetylated histone tails. CpG methylation induces histone deacetylation, chromatin remodeling and gene silencing through a transcription repressor complex that includes SMRT (Silencing Mediator of Retinoid and Thyroid Receptors), mSin3a, RbAp46/48 and the two histone deacetylases HDAC1 and HDAC2 formed around mSin3a. This complex is assembled by interaction of mSin3a with the methyl-binding protein MeCP2 and SAP18/30 (Sin3-Associated Polypeptides 18/30), which are found associated with large protein complexes such as the NURD complex (Methyl-CpG Binding Domain proteins: MBD2, MBD3). MECP2 acts as a shuttle interlocking DNA methylation and core histone deacetylation in inducing gene silencing. This methyl-binding protein tethers the repression multiprotein complex that includes the corepressor protein, mSin3a, HDAC1 and HDAC2 (Ref.3). The deacetylase activity, which accompanies the MeCP2-bound mSin3a render the promoter of the gene inaccessible to TFs by deacetylating histone H3 and H4. To reverse such a silenced status of a gene, demethylation takes place and an activating complex, which carries the capacity of acetylating histones H3 and H4, replaces the repression complex. This modification of core histones results in a chromatin structure, which is accessible to TFs. Alternatively, the methyl-directed repression can be alleviated by a methylation-overriding effect that is exerted by a strong activation complex ultimately resulting in effective acetylation of histones H3 and H4. In addition to MBD, a TRD (Transcriptional Repressor Domain) overlaps with a region that interacts directly with the corepressor mSin3a (Ref.4). HDAC1 and HDAC2 and chromatin-remodeling activities (Mi-2 and mSin3a) within these complexes result in alterations in chromatin structure, producing chromatin that is refractory to transcriptional activation.
The repression mechanism is significantly different when the number of methylated sites is increased and reaches the threshold that leads to diffusion of gene silencing on the DNA fiber. Hypomethylation contributes to chromosomal instability and possibly to increased expression of some proto-oncogenes. CpG island methylation is capable of silencing tumor suppressor genes and also increases the possibility of mutations, which can occur frequently in these regions. DNA methylation at the 5-position of cytosine within CpG dinucleotides in mammals is essential for important functions, such as cell differentiation, imprinting and X-inactivation. Several genetic diseases are caused by defects within the methylation machinery, like the Rett Syndrome, Fragile X Syndrome and ICF (Immunodeficiency-Centromeric Instability-Facial Anomalies Syndrome).
References:
1.Repression: targeting the heart of the matter.
Maldonado E, Hampsey M, Reinberg D.
Cell. 1999 Nov 24; 99(5): 455-8. Review. No abstract available.
2.Molecular mechanisms of gene silencing mediated by DNA methylation.
Curradi M, Izzo A, Badaracco G, Landsberger N.
Mol Cell Biol. 2002 May; 22(9): 3157-73.
3.Linker histone binding and displacement: versatile mechanism for transcriptional regulation.
Zlatanova J, Caiafa P, Van Holde K.
FASEB J. 2000 Sep; 14(12): 1697-704. Review.
4.CpG methylation, chromatin structure and gene silencing-a three-way connection.
Razin A.
EMBO J. 1998 Sep 1; 17(17): 4905-8. Review.
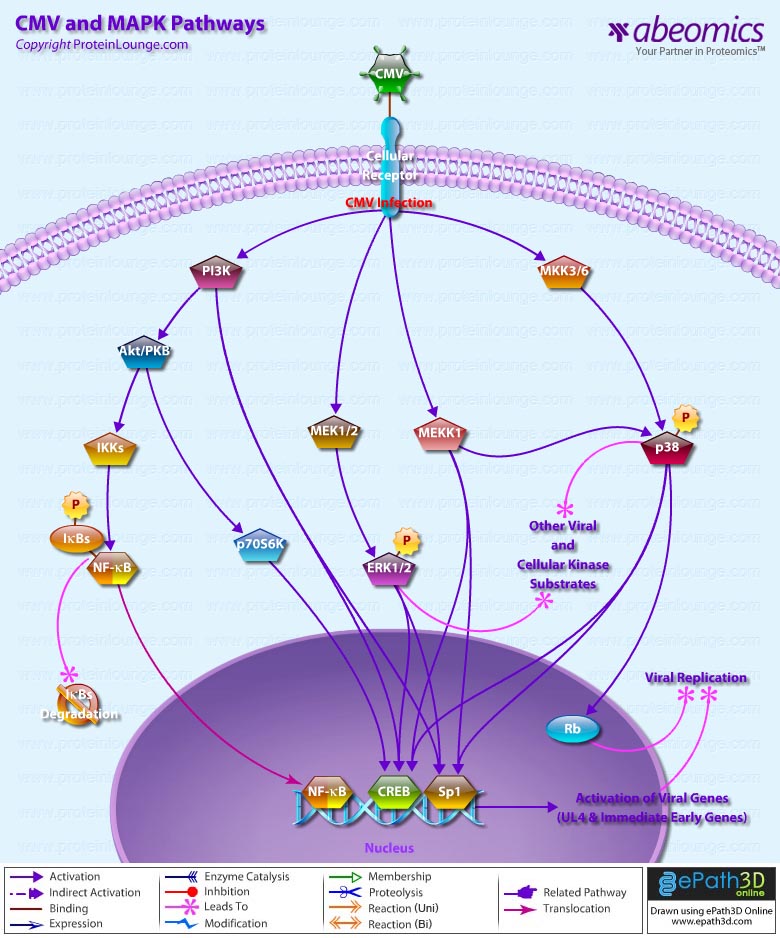
Signal transduction is a common process used by an extensive array of biological ligands to modulate various host cell processes such as growth, differentiation, and proliferation. Cells respond to CMV Cytomegalovirus) by invoking a cascade of biological and physiological responses resulting in signal transduction and regulation of cellular gene expression, including induction of genes in the IFN (Interferons)-responsive family (Ref.1). CMV is a herpes virus that infects most cell types and establishes latency in leukocytes. A CMV infection is normally subclinical but can be fatal in immunocompromised individuals or if the infection is acquired in utero.
One of the major components of the CMV virion is the Glycoprotein GB, and a significant proportion of the virus-host interactions are mediated through GB. CMV stimulates a large number of typical proinflammatory signaling events, including nuclear translocation of NF-kappaB (Ref.2). The first step in CMV infection is attachment to its cellular target; however, the receptors for CMV have not yet been delineated. Heparin sulfate and CD13 have been suggested to be involved in CMV attachment (Ref.3). One of the early cellular responses to HCMV (Human CMV) infection is production of IP3 (Inositol 1,4,5-triphosphate) and DAG (1,2-Diacylglycerol) from PI3K (Phosphatidylinositol 3-Kinases) by phosphorylation at the D-3 position. Once phosphorylated at the D-3 position, these lipids serve as second messengers and are able to regulate phosphorylation of a number of kinases, including Akt1/PKB (Protein Kinase-B), cAMP (cyclic Adenosine 3′, 5’-Monophosphate)-dependent PKA (Protein Kinase-A), some isoforms of PKC (Protein Kinase-C), p70S6K and p85S6K (Ribosomal S6 Kinases), respectively (Ref.4). In addition, the HCMV particle in the host cell carry phosphatase activity and activate the membrane-proximal PLC (Phospholipases-C) and A2 as well as the prototypical MAPK (Mitogen-Activated Protein Kinase) pathway. The MAPK signal transduction cascade consists of a three-component module consisting of MAPK, MEK (MAP/ERK Kinase), and MEKK (MAP/ERK Kinase Kinase), which are conserved from yeasts to humans (Ref.5). MAPK activation by CMV leads to activation of transcription of viral genes, increasing the production of viral gene products. The MEKK1 regulates the immediate early promoter indirectly through downstream kinase signaling and directly through activation of NF-kappaB (Nuclear Factor-kappaB). The mechanism of activation of NF-kappaB involves the liberation of transcription factor from the inhibitory subunit I-kappaB-Alpha (Inhibitor of Kappa Light Chain Gene Enhancer in B-Cells-Alpha) and is mediated by a GB-dependent mechanism through IKKs (I-KappaB Kinases). The early activation of NF-kappaB is amplified by other mechanisms later during infection. This second wave of NF-kappaB activation relies on NF-kappaB-dependent activation of the CMV major immediate-early promoter. MAPK pathways activated by CMV converge on increased transcription of viral genes and increased replication of the viral genome. ERK1/2 (Extracellular Signal-Regulated Kinases) and p38 immediately follow infection that regulates the activity of viral genes by cellular transcription factors acting through the basal transcription elements and viral UL4 promoter located upstream. Another target of prolonged p38 activation during infection is Rb (Retinoblastoma), contributing to viral replication (Ref.6). The classical binding partner for Rb is the E2F family of transcription factors, and hyperphosphorylation of pRb is necessary for relieving pRb-mediated suppression of E2F, which results in cell cycle progression past the G1/S-phase transition point. However, the HCMV IE2-86 protein binds to pRb and alleviates pRb-mediated suppression of E2F transactivation function.
Cellular proto-oncogenes such as c-Fos, c-Jun, and c-Myc, which are upregulated in response to mitogenic stimuli, are also stimulated by HCMV infection. Furthermore, TPA (12-O Tetradecanoylphorbol-13-Acetate), which stimulates the ERK pathway activate the HCMV MIEP (Ref.7). The MAPKs also activate viral transactivators, which are important in the activation of early viral promoters. For example, ERK phosphorylates both IE72 and IE86 proteins in vitro and in vivo. In addition, MEK1/2 inhibitors UO126 and PD98059 repress phosphorylation of IE72 and IE86 proteins in infected cells without decreasing the level of IE proteins (Ref.5). Cooperation of IE1-72, IE2-55, and IE2-86 proteins with the cellular transcription factor Sp1 (Selective promoter factor-1) up-regulates the promoters for the NF-kappaB subunits p65 and p105/p50. HCMV has also been reported to activate IRF3 (Interferon Regulatory Factor-3) through a rapid de novo protein synthesis-independent mechanism.
HCMV is a widespread human pathogen that does not cause significant clinical manifestations in healthy individuals. Following primary infection HCMV persists for life as a latent infection with periodic asymptomatic excretion of virus in saliva, breast milk, urine, semen and cervical secretions. The major neutralization epitopes of HCMV are located on its glycoprotein, GB. CMV in immunocompetent individuals is etiologically associated with infectious mononucleosis as well as other disease presentations and malignancies (Ref.8) and if left untreated, can be fatal. In addition, it is a leading cause of certain types of birth defects. Individuals suffering from diseases caused by HCMV are currently treated with chemical compounds, such as ganciclovir and phosphocarnet, which block the viral lytic life cycle by inhibiting viral DNA replication. However, the substantial toxicity of these drugs and the emergence of drug-resistant strains of HCMV indicate that better antiviral compounds are needed (Ref.9).
References:
1.Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway.
Boyle KA, Pietropaolo RL, Compton T
Mol Cell Biol. 1999 May; 19(5): 3607-13
2.The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-kappaB during infection
Yurochko AD, Hwang ES, Rasmussen L, Keay S, Pereira L, Huang ES
J Virol. 1997 Jul; 71(7): 5051-9
3.Productive cytomegalovirus (CMV) infection exclusively in CD13-positive peripheral blood mononuclear cells from CMV-infected individuals: implications for prevention of CMV transmission
Larsson S, Soderberg-Naucler C, Moller E
Transplantation. 1998 Feb 15; 65(3): 411-5
4.The PI3K-PDK1 connection: more than just a road to PKB
Vanhaesebroeck B, Alessi DR
Biochem J. 2000 Mar 15; 346 Pt 3:561-76. Review
5.Role of regulatory elements and the MAPK/ERK or p38 MAPK pathways for activation of human cytomegalovirus gene expression
Chen J, Stinski MF
J Virol. 2002 May; 76(10): 4873-85
6.A protein kinase involved in the regulation of inflammatory cytokine biosynthesis
Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW, et al
Nature. 1994 Dec 22-29; 372(6508): 739-46
7.Extracellular signal-regulated kinase activity is sustained early during human cytomegalovirus infection
Rodems SM, Spector DH
J Virol. 1998 Nov; 72(11): 9173-80
8.Epstein-Barr virus latent infection in vivo
Steven NM
Rev Med Virol. 1997 Jul; 7(2): 97-106
9.Histone acetylation and reactivation of Epstein-Barr virus from latency
Jenkins PJ, Binne UK, Farrell PJ
J Virol. 2000 Jan; 74(2): 710-20
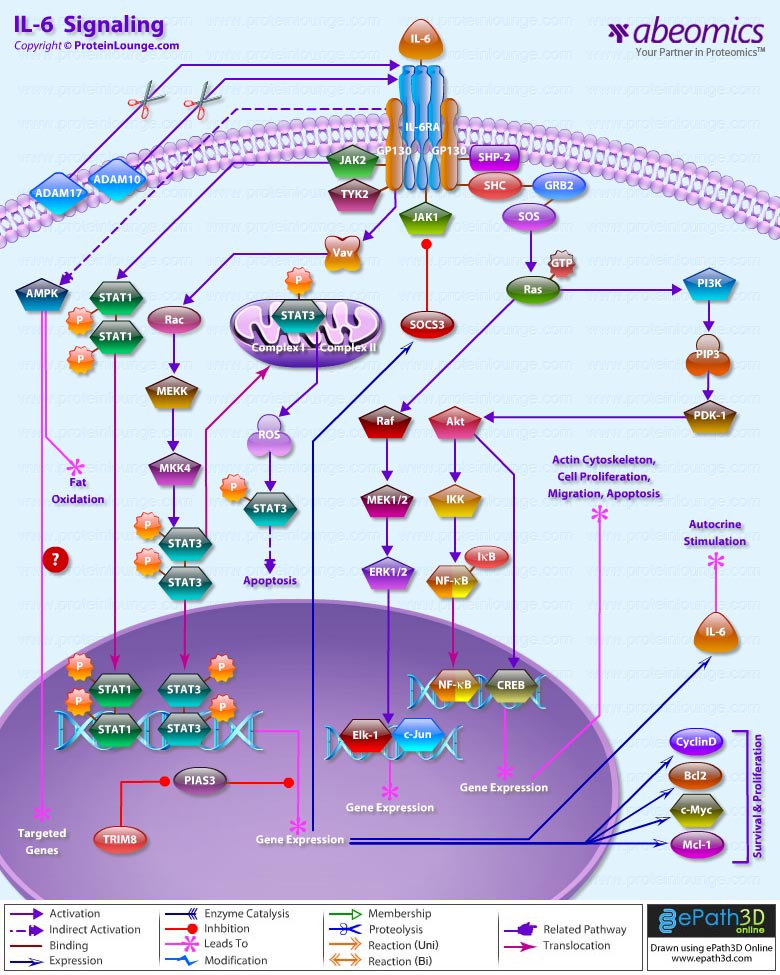
IL(Interleukin)-6 is a pleiotropic cytokine that not only affects the immune system, but also acts in other biological systems and many physiological events in various organs including the inflammation, hematopoiesis, and oncogenesis by regulating cell growth, gene activation, proliferation, survival, and differentiation. This proteinsignals through a receptor composed of two different subunits, an alpha subunit that produces ligand specificity and GP (Glycoprotein) 130, a receptor subunit shared in common with other cytokines in the IL-6 family. Binding of IL-6 to its receptor initiates cellular events including activation of JAK (Janus Kinase) kinases and activation of Ras-mediated signaling. Activated JAK kinases phosphorylate and activate STAT transcription factors, particularly STAT3 (Signal Transducers and Activators of Transcription-3) and SHP2 [SH2 (Src Homology-2) Domain-containing Tyrosine Phosphatase] (Ref.1). Phosphorylated STAT3 then forms a dimer and translocates into the nucleus to activate transcription of genes containing STAT3 response elements. STAT3 is essential for GP130-mediated cell survival and G1 to S cell-cycle-transition signals. Both c-Myc and Pim have been identified as target genes of STAT3 and together can compensate for STAT3 in cell survival and cell-cycle transition. SHP2 links cytokine receptor to the Ras/MAP (Mitogen-Activated Protein) kinase pathway and is essential for mitogenic activity (Ref.2&3).
The Ras-mediated pathway,acting through SHC, GRB2 (Growth Factor Receptor Bound protein-2) and SOS1 (Son of Sevenless-1) upstream and activating MAP kinases downstream, activates transcription factors such as Elk1 and NF-IL-6 (C/EBP-Beta) that can act through their own cognate response elements in the genome. These factors and other transcription factors like Activating Protein-1 and SRF (Serum Response Factor) that respond to many different signaling pathways come together to regulate a variety of complex promoters and enhancers that respond to IL-6 and other signaling factors (Ref.4).In addition to JAK/STAT and Ras/MAP kinase pathways, IL-6 also activates PI3K (Phosphoinositide-3 Kinase). The PI3K/Akt/NF-KappaB cascade activated by IL-6, functions cooperatively to achieve the maximal anti-apoptotic effect of IL-6 against TGF-Beta(Transforming Growth Factor-Beta). The anti-apoptotic mechanism of PI3K/Akt is attributed to phosphorylation of the BCL2 (B-Cell Leukemia-2) family member BAD (BCL2 Associated Death Promoter) by Akt. The phosphorylated BAD is then associated with 14-3-3, which sequesters BAD from BCLXL, thereby promoting cell survival. Regulating the BCL2 family member is also considered as one of the anti-apoptotic mechanisms of STAT3, which was reported to be capable of inducing BCL2 in pro-B cells. Thus, both anti-apoptotic signaling pathways transduced by IL-6 are likely to converge to BCL2 family members, which could act upstream of Caspase3. IL-6 also blocks the TGF-Beta induced activation of Caspase3. In addition to induction of BCL2, STAT3 can directly up-regulate the transcription of p21, which is implicated in the anti-apoptosis. The termination of the IL-6-type cytokine signaling is through the action of tyrosine phosphatases, proteasome, and JAK kinase inhibitors SOCS (Suppressor of Cytokine Signaling), PIAS (Protein Inhibitors of Activated STATs), and internalization of the cytokine receptors via GP130. One of the major actions of IL-6 is the transcriptional activation of APP (Acute-Phase Plasma Proteins) genes in liver cells. SHP2 acts as a negative regulator of the JAK/STAT signaling in part by downregulating JAK activity, thereby indirectly moderating the induction of STAT3-dependent APP genes. IL-6 stimulates several types of Leukocytes, and the production of Acute Phase Proteins in the Liver. It is particularly important in inducing B-Cells to differentiate into Antibody Forming Cells (Plasma Cells).IL-6 is released into the circulation, where it works in a hormone-like fashion to induce lipolysis and fat oxidation. In more recent experiments, it has been shown that IL-6 infusion increases glucose disposal during a hyperinsulinaemic euglycaemic clamp in healthy humans. IL-6 treatment of myotubes increases fatty acid oxidation, basal and insulin-stimulated glucose uptake and translocation of GLUT4 to the plasma membrane. Furthermore, IL-6 rapidly and markedly increases AMPK (AMP-activated protein kinase) and the metabolic effects of IL-6 were abrogated in AMPK dominant negative-infected cells. Finally, IL-6 mediates anti-inflammatory effects by stimulating the production of anti-inflammatory cytokines and by suppressing TNFalpha production. We suggest that IL-6 and other muscle-derived cytokines (myokines) may play a role in defending Type 2 diabetes(Ref.4,5& 6)
References:
1.Interleukin-6-induced JAK2/STAT3 signaling pathway in endothelial cells is suppressed by hemodynamic flow. Ni CW, Hsieh HJ, Chao YJ, Wang DL. Am J Physiol Cell Physiol. 2004 Sep;287(3):C771-80. Epub 2004 May 19.
2.Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Chem Biol. 2006 Nov;13(11):1235-42.
3.Modulation of dendritic cell differentiation by HLA-G and ILT4 requires the IL-6–STAT3 signaling pathway. Liang S, Ristich V, Arase H, Dausset J, Carosella ED, Horuzsko A. Proc Natl Acad Sci U S A. 2008 Jun 17;105(24):8357-62. Epub 2008 Jun 11.
4.Molecular regulation of cardiac hypertrophy. Barry SP, Davidson SM, Townsend PA. Int J Biochem Cell Biol. 2008;40(10):2023-39. Epub 2008 Feb 26. Review.
5.The enigma of the role of protein inhibitor of activated STAT3 (PIAS3) in the immune response. Yagil Z, Nechushtan H, Kay G, Yang CM, Kemeny DM, Razin E. Trends Immunol. 2010 May;31(5):199-204. Epub 2010 Feb 22. Review. 6.IL-6 signalling in exercise and disease. Pedersen BK.
Biochem Soc Trans. 2007 Nov;35(Pt 5):1295-7. Review.
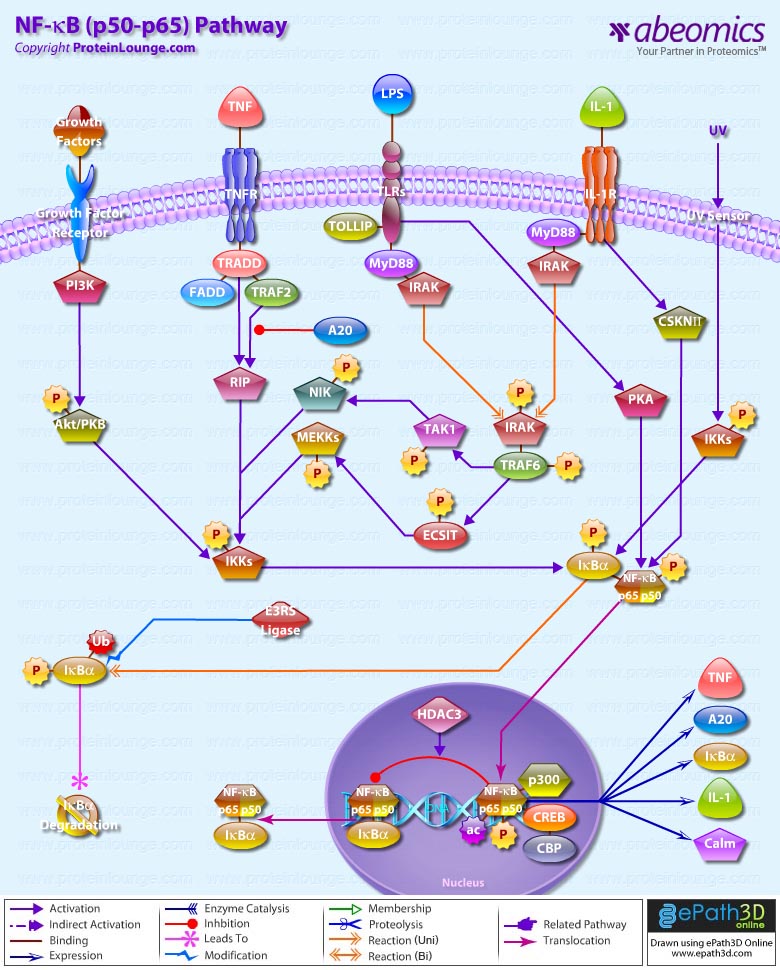
Survival of an organism is dependent on its ability to rapidly and effectively respond to adverse changes in its environment. Eukaryotic cells possess a number of distinct signal transduction pathways that couple environmental stimuli to specific changes of gene expression. One such pathway is the transcription factor NF-KappaB (Nuclear Factor-KappaB), which is implicated in the regulation of many genes that code for mediators of apoptosis, viral replication, tumorigenesis, various autoimmune diseases and inflammatory responses (Ref.1). NF-KappaB is composed of homo- and heterodimeric complexes of members of the Rel (NF-KappaB) family: p50, p65 (RelA), c-Rel, p52 and RelB. The most common and best-characterized form of NF-KappaB is the p65/p50 heterodimer. Each dimer combination exhibits differences in DNA binding affinity and transactivation potential. The activation of NF-KappaB is thought to be part of a stress response as it is activated by a variety of stimuli that include growth factors, cytokines, such as TNF (Tumor Necrosis Factor) and IL-1 (Interleukin-1) lymphokines, components of bacterial cell walls, such as LPS (Lipopolysaccharide), UV, pharmacological agents, and stress (Ref.2).
Depending on the stimulus, the mechanism of activation involves overlapping and nonoverlapping steps. Among all the stimuli, perhaps the most is known about the mechanism by which TNF activates NF-KappaB. In its inactive form, NF-KappaB is sequestered in the cytoplasm, bound by members of the I-KappaB family of inhibitor proteins. The activity of NF-KappaB is regulated by inhibitory proteins, which include I-KappaB-Alpha, I-KappaB-Beta, I-KappaB-Gamma, I-KappaB-Epsilon, BCL3, p105, and p100. I-KappaB proteins are phosphorylated by I-KappaB Kinase complex consisting of at least three proteins, IKK-Alpha, IKK-Beta, and IKK-Gamma (Ref.3).
NF-KappaB pathway involves the interaction of the ligand with its receptor at the cell surface (TNFR), which then recruits a protein called TRADD (TNF Receptor-Associated Death Domain). This protein binds to TRAF2 (TNF Receptor-Associated Factor-2), which activates RIP (Receptor-Interacting Protein). RIP interacts with MEKK (Mitogen-Activated Protein Kinase Kinase) and NIK (NF-KappaB-Inducing Kinase) to phosphorylate and activate the IKK (I-KappaB-Alpha kinase complex). The IKK complex phosphorylates I-KappaB-Alpha, which leads to ubiquitination and then leads to the degradation of I-KappaB-Alpha by the proteosome, resulting in the translocation of NF-KappaB to the nucleus. In the nucleus it binds to its consensus sequence (5′-GGGACTTTC-3′) and positively regulates the transcription of genes involved in immune and inflammatory responses, cell growth control, and apoptosis. Genes encoding cytokines, cytokine receptors, cell adhesion molecules, chemoattractant proteins, and growth regulators are positively regulated by NF-KappaB (Ref.4). TRAF2 also interacts with A20, a zinc finger protein whose expression is induced by agents that activate NF-KappaB. A20 functions to block TRAF2-mediated NF-KappaB activation
IL-1 has similar downstream affects through NF-KappaB, including immunoregulation, proinflammatory, and hematopoietic activities. IL-1 induced signaling is mediated through association of IL-1 receptors with adaptor proteins, such as IRAK (IL-1 Receptor-Associated Kinases) and MyD88 (Myeloid Differentiation Primary Response Gene-88). Activation of NF-KappaB by bacterial LPS promotes the upregulation of proinflammatory cytokines. Following recognition of LPS, the adapter protein MyD88 is recruited to the cytoplasmic domain of TLR4 (Toll Like Receptor-4). MyD88 contains a highly conserved DD (Death Domain) that facilitates its interaction with another DD-containing signaling molecule, IRAK. Following recruitment to MyD88, IRAK undergoes rapid autophosphorylation and dissociation from the signaling complex. Phosphorylated IRAK subsequently interacts with TRAF6, initiating the activation of a kinase cascade involving NIK and IKK. Activation of this cascade culminates in the phosphorylation and degradation of the I-KappaB, enabling NF-KappaB to translocate to the nucleus and promote new gene expression (Ref.5). NF-KappaB-dependent gene expression involves a growing family of proteins termed transcriptional coactivators that probably function by facilitating or bridging the sequence-specific activators to the basal transcriptional machinery and altering chromatin structure. The p65 component of NF-KappaB binds to the coactivator CBP (cyclic AMP response element binding protein [CREB]-binding protein) and its structural homolog p300. Phosphorylation of p65 by PKA (Protein Kinase-A) stimulates NF-KappaB-dependent gene expression by enhancing p65 association with CBP. Recent observations have shown that levels of the CBP homolog, p300, are limiting relative to those of p65 and that competition for CBP may regulate p65 transactivation. Deacetylation of p65 through specific interactions with HDAC3 (Histone Deacetylase-3) promotes effective binding to newly synthesized I-kappaB-Alpha, which subsequently leads to I-kappaB-Alpha dependent nuclear export. Overexpression of the p110 catalytic subunit of PI3K (Phosphoinositide-3 Kinase) also induces p65 -mediated transactivation and that the specific PI3K inhibitor LY294002 represses this process. Additionally, the expression of a constitutively activated form of either p110 or the PI3K-activated protein kinase Akt also induces p65/RelA-mediated transactivation. NF-KappaB pathways provide many targets for developing specific drugs to treat inflammatory diseases. Inappropriate activation of NF-KappaB has been linked to inflammatory events associated with autoimmune arthritis, asthma, septic shock, lung fibrosis, glomerulonephritis, atherosclerosis, and AIDS.
References:
1.The NF-kappa B and I kappa B proteins: new discoveries and insights.
Baldwin AS Jr.
Annu Rev Immunol. 1996;14:649-83.
2.Two pathways to NF-KappaB.
Joel L. Pomerantz and David Baltimore.
Molecular Cell, Vol. 10, 693-701, October 2002June 2002, Volume 16, Number 6, Pages 1053-1068.
3.I-kappa-B–NF-kappa-B structures: at the interface of inflammation control.
Baeuerle, P. A.
Cell 95: 729-731, 1998.
4.NF-KappaB signaling: Many Roads Lead To Madrid.
Vishva Dixit and Tak. W. Mak.
Cell, Vol. 111, 615-619, November 27, 2002.
5.The zinc finger protein A20 inhibits TNF-induced NF-KappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-KappaB-inhibiting protein ABIN.
Heyninck K, De Valck D, Vanden Berghe W et al.
J Cell Biol 1999 Jun 28;145(7):1471-82.
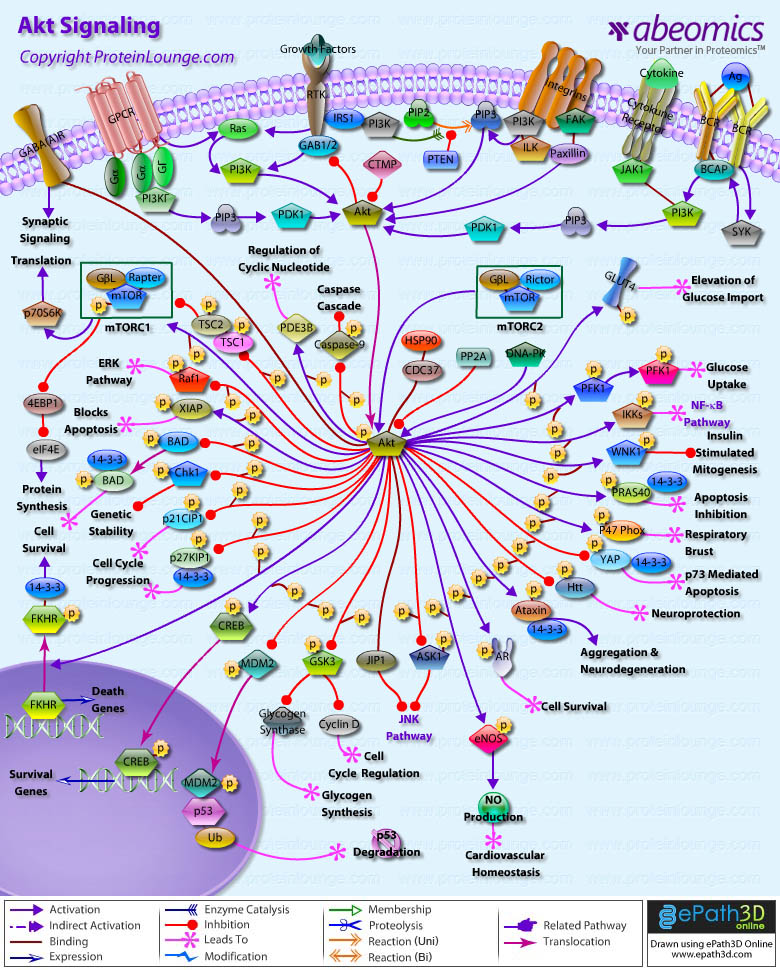
AKT/PKB Pathway is an evolutionarily conserved serine/threonine kinase Pathway involved in a wide variety of cellular functions, including proliferation, cell survival, differentiation, glucose mobilization, homeostasis, cell migration, and apoptosis. Three isoforms, AKT1,AKT2, and AKT3, are expressed in mammals (Ref.1 & 2). Akt is a central node in cell signaling downstream of growth factors, cytokines, and other cellular stimuli. Aberrant loss or gain of Akt activation underlies the pathophysiological properties of a variety of complex diseases, including type-2 diabetes and cancer (Ref.3). All three isoforms of Akt share a common structure of three domains. The N-terminus of the protein is a PH (Pleckstrin Homology) domain, which interacts with membrane lipid products such as PIP2 (Phosphatidylinositol-3,4-Bisphosphate) and PIP3 (Phosphatidylinositol-3,4,5-Triphosphate). The PH domain is approximately 100 amino acids and plays a role in recognition by upstream kinases and membrane translocation of Akt. The center region of the protein is the Kinase domain, which has high similarity to other kinases. This domain contains a conserved threonine residue, which needs to be phosphorylated in order to activate Akt. The approximately 40 amino acids at the C-terminus of the protein form a regulatory domain that contains a proline rich region and a hydrophobic motif with a conserved sequence of FXX (F/Y)(S/T)(Y/F). In mammals, this hydrophobic motif is FPQFSY. The serine or threonine residue in this motif must also be phosphorylated to activate Kinase activity of Akt. This is also a conserved residue (Ref.4).
Activation of Akt can begin with several events, mainly the binding of a Ligand to a Receptor in the cell membrane. Most common Ligands activating Akt include Growth factors, Cytokines, Mitogens and Hormones. Insulin and a variety of Growth factors bind to RTK (Receptor Tyrosine Kinase) and cause autophosphorylation of tyrosine residues on the intracellular domain of the receptor. PI3K (Phosphoinositol 3-Kinase) is recruited to the phosphotyrosine residues (consensus sequence pYXXM) via SH2 domains in the regulatory domain (p85), and is therefore targeted to the inner cell membrane. Binding of the p85 subunit of PI3K to the phosphorylated RTK leads to conformational changes in the catalytic domain of PI3K (p110) and consequent kinase activation. PI3K can be activated by Ras. Insulin can also activate PI3K via IRS1 (Insulin Receptor Substrate-1). GPCR (G-Protein-Coupled Receptor) also activates PI3K through GN-Beta (Guanine Nucleotide-Binding Protein-Beta) and GN-Gamma (Guanine Nucleotide-Binding Protein-Gamma) subunits of G-proteins. Cytokines can also activate PI3K via JAK1 (Janus Kinase-1). In B-Cells, PI3K is activated by BCR (B-Cell Receptor) via SYK (Spleen Tyrosine Kinase) and BCAP (B-Cell Receptor Associated Protein). PI3K then phosphorylates membrane bound PIP2 to generate PIP3. The binding of PIP3 to the PH domain anchors Akt to the plasma membrane and allows its phosphorylation and activation by PDK1 (Phosphoinositide-Dependent Kinase-1). DNA-PK, CDC37 (Cell Division Cycle-37), HSP90 (Heat Shock Protein-90KD) and PKCƒ{Beta (Protein Kinase-C-Beta) are also reported to phosphorylate Akt. Integrins also activates Akt via FAK (Focal Adhesion Kinase), Paxillin and ILK (Integrin-Linked Kinase). Akt can also be activated in response to a variety of cellular stress, such as heat shock, administration of ultra violet light, ischemia (a decrease in blood supply), hypoxia (oxygen deficiency), hypoglycemia (abnormally low level of glucose in the blood) and oxidative stress. The activity of Akt is negatively regulated by PTEN (Phosphatase and Tensin Homolog), SHIP (SH2-Containing Inositol Phosphatase) and CTMP (Carboxyl-Terminal Modulator Protein) (Ref.5, 6 & 7).
The actions of Akt in the cell are numerous and diverse, but all result in anti-apoptosis, or pro-cell proliferation effects. These physiological roles of Akt include involvement in metabolism, protein synthesis, apoptosis pathways, transcription factor regulation and the cell cycle. Akt exerts its effects in the cell by phosphorylating a variety of downstream substrates. The downstream targets of Akt include BAD (BCL2 Antagonist of Cell Death), Caspase9, FKHR (Forkhead Transcriptional Factor), GLUTs (Glucose Transporters), eNOS (Nitric Oxide Synthase), PFK2 (6-Phosphofructo-2-Kinase), PFK1(6-Phosphofructo-Kinase), mTOR (Mammalian Target of Rapamycin), IKK (I-KappaB Kinase), NF-KappaB (Nuclear Factor-KappaB), GSK3 (Glycogen Synthase Kinase-3), WNK1(WNK Lysine deficient Protein Kinase-1), PRAS40 (Proline-Rich Akt Substrate 40 kDa), p47Phox, YAP (Yes-Associated Protein-1), Htt (Huntingtin), Ataxin, AR (Androgen Receptor), ASK1 (Apoptosis Signal-Regulating Kinase-1), MDM2 (Mouse Double Minute-2), CREB (cAMP Response Element-Binding Protein), p21CIP1 (Cyclin Dependent Kinase Inhibitor-p21), p27KIP1 (Cyclin Dependent Kinase Inhibitor-p27) , Chk1 (Cell Cycle Checkpoint Kinase-1), XIAP (X-Linked Inhibitor of Apoptosis Protein), Raf1 (v-Raf1 Murine Leukemia Viral Oncogene Homolog-1), PDE3B (Phosphodiesterase 3B cGMP-Inhibited), TSC (Tuberous Sclerosis Gene) and GABA(A)R (Gamma-aminobutyric Acid Receptor-A) (Ref.8).
Akt inhibits apoptosis by phosphorylating the BAD component of the BAD/BclXL (Bcl2 Related Protein Long Isoform) complex. Phosphorylated BAD binds to 14-3-3 causing dissociation of the BAD/BclXL complex and allowing cell survival. Akt activates IKK, which ultimately leads to NF-KappaB activation and cell survival. Other direct targets of Akt are members of the FKHRL1 (Forkhead-Related Family of Mammalian Transcription Factor-1). In the presence of survival factors, Akt1 phosphorylates FKHRL1, leading to the association of FKHRL1 with 14-3-3 proteins and its retention in the cytoplasm. Survival factor withdrawal leads to FKHRL1 dephosphorylation, nuclear translocation, and target gene activation. Within the nucleus, FKHRL1 most likely triggers apoptosis by inducing the expression of genes that are critical for cell death, such as the TNFSF6 (Tumor Necrosis Factor Ligand Superfamily Member-6) gene. Another notable substrate of Akt is the death protease Caspase9. Phosphorylation of Caspase9 decreases apoptosis by directly inhibiting the protease activity. Akt also activates TERT (Telomere Reverse Transcriptase), which is responsible for telomere maintenance and DNA stability. Akt has been linked to angiogenesis, through the activation of eNOS, which influences long-term blood vessel growth. Akt can regulate several levels of Glucose metabolism. It enhances Glucose-uptake in Insulin-responsive tissues by inducing the expression of GLUT1 and GLUT3 and the translocation of GLUT4 to the plasma membrane; the GLUTs transport glucose into the cell. Akt also activates Glycogen synthesis by phosphorylating and inactivating GSK3, which leads to the activation of Glycogen Synthase and CyclinD1. Akt phosphorylates PDE3B on Ser273. This activates PDE3B and results in regulation of intracellular levels of cyclic nucleotides in response to Insulin. Akt induces glycolysis through the phosphorylation and activation PFK2, which in turn activates PFK1. These enzymes convert Fructose-6-Phosphate into Fructose-1, 6-Bisphosphate, a key step in Glucose metabolism. Akt may also be involved in activation of the nutrient-dependent Thr/Ser kinase, mTOR.Activation of mTOR results in the phosphorylation of ribosomal protein S6 kinase, p70S6K. Akt also phosphorylates the two tumor suppressor genes TSC1 and TSC2, which are negative regulators of the mTOR-S6K pathway. Phosphorylation of TSC1 and TSC2 results in suppression of their inhibitory activity and may also target the proteins for degradation. Activation of mTOR also results in phosphorylation and inactivation of eIF4EBP (Eukaryotic Initiation Factor-4E Binding Protein), an inhibitor of the translation initiation factor eIF4E. Nonphosphorylated PHASI binds to eIF4E (Eukaryotic Initiation Factor-4E) and inhibits protein synthesis. Akt also phosphorylates GAB2 (GRB2-Associated Binding Protein-2) on Ser159. Phosphorylation of Ser159 on Gab2 by Akt/PKB appears to negatively regulate GAB2 tyrosine phosphorylation by the ErbB receptor tyrosine kinases, although the underlying mechanism has not been solved (Ref.9).
The transcription factor CREB is directly phosphorylated at Ser133 by Akt. This causes an increased affinity of CREB for its co-activator protein, CRB (Crumbs). The heterodimer, now an active transcription factor, promotes transcription of genes that contain CREs (cAMP responsive elements) in their promoter, such as the anti-apoptotic genes Bcl2 and Mcl1. Akt also phosphorylates AR at two serine residues, Ser210 and Ser270, which causes a decrease in AR activity on the p21 promoter. In addition to causing cell cycle progression, this also results in apoptosis inhibition in certain cell types, through other actions of AR. YAP is another transcription factor that is phosphorylated by Akt, and is of importance because it does not contain an Akt consensus sequence. Akt phosphorylates Ser127 on YAP, which causes association with 14-3-3 proteins, nuclear export and cytoplasmic localization. Akt has also been shown to phosphorylate p21 directly, on Thr145. p21 is a member of the Cip/Kip family of CDK inhibitors that arrest the cell cycle and therefore limit cell proliferation. p21 can also promote cell cycle progression, via mediating the assembly and activity of cyclin D1-CDK4/6 complexes. P27 is another cyclin-dependent kinase inhibitor, of the Kip family. P27 inhibits CDK2 and CDK4/6 complexes, which is located in the nuclear localization signal. NLS targets protein to nucleus via nuclear import machinery, and phosphorylation in this region of p27 results in nuclear exclusion. 14-3-3 proteins bind phosphorylated p27 and cause active export from nucleus. Without p27 in the nucleus, the cyclin-CDK complexes form and promote cell cycle progression. Akt also phosphorylates MDM2. MDM2 is phosphorylated at many sites, only two of which have been identified. Ser166 is phosphorylated by Akt. Akt phosphorylation of MDM2 allows its entry into the nucleus where it targets p53 for degradation (Ref.10, 11 & 12).
PRAS40 is a 40 kDa substrate of AKT. Activated AKT phosphorylates PRAS40 on threonine 246, enabling PRAS40 to bind to 14-3-3. AKT and PRAS40 are components of the PI3K pathway. This pathway plays a role in glucose uptake, cell growth, and apoptosis inhibition. The precise function of PRAS40 is not yet known; however, it has been hypothesized that PRAS40 interacts with SH3 and WW domain containing proteins, and may change the function of these proteins. Akt phosphorylates, both in vitro and in vivo, the GABA(A)R, the principal receptor mediating fast inhibitory synaptic transmission in the mammalian brain. Akt- mediated phosphorylation increases the number of GABA(A)Rs on the plasma membrane surface, thereby increasing the receptor-mediated synaptic transmission in neurons. XIAP is a physiological substrate of Akt. Akt interacts with and phosphorylates XIAP at serine 87. Phosphorylation of XIAP by Akt inhibits both its autoubiquitination and cisplatin-induced ubiquitination. These effects reduce XIAP degradation and the increased levels of XIAP are associated with decreased cisplatin-stimulated Caspase3 activity and programmed cell death. Htt is also a substrate of Akt and phosphorylation of Htt by Akt is crucial to mediate the neuroprotective effects of IGF1 (Insulin-Like Growth Factor-I). WNK1 is a physiologically relevant target of Insulin signaling through PI3K and Akt and functions as a negative regulator of Insulin-stimulated mitogenesis (Ref.13, 14 & 15).
Akt also phosphorylates Ataxin1 and modulate neurodegeration.14-3-3 protein mediates the neurotoxicity of Ataxin1 by binding to and stabilizing Ataxin1, thereby slowing its normal degradation. Akt also decreases ASK1 kinase activity by phosphorylating a consensus Akt site at serine 83 of ASK1. Akt also interacts with the JIP1 (JNK Interacting Protein-1) scaffold and inhibits the ability of JIP1 to form active JNK signaling complexes. The binding of Akt to JIP1 is isoform specific; Akt1 but not Akt2 interacts with JIP1. Thus, Akt can inhibit one or more steps within the JNK signaling pathway, depending on the complement of components that form the functional JNK signaling module. Akt mediates PI3K-dependent p47Phox phosphorylation, which contributes to respiratory burst activity in human neutrophils. AKT impair Chk1 through phosphorylation, ubiquitination, and reduced nuclear localization to promote genomic instability in tumor cells. Akt and its upstream regulators are deregulated in a wide range of solid tumors and hematologic malignancies, hence the Akt pathway is considered a key determinant of biologic aggressiveness of these tumors, and a major potential target for novel anti-cancer therapies (Ref.16 & 17).
References:
1.Crowell JA, Steele VE, Fay JR.
Targeting the AKT protein kinase for cancer chemoprevention.
Mol Cancer Ther. 2007 Aug;6(8):2139-48. Review.
2.Cenni V, Bavelloni A, Beretti F, Tagliavini F, Manzoli L, Lattanzi G, Maraldi NM, Cocco L, Marmiroli S.
Ankrd2/ARPP is a novel Akt2 specific substrate and regulates myogenic differentiation upon cellular exposure to H(2)O(2).
Mol Biol Cell. 2011 Aug 15;22(16):2946-56. doi: 10.1091/mbc.E10-11-0928. Epub 2011 Jul 7.
3.Manning BD, Cantley LC.
AKT/PKB signaling: navigating downstream.
Cell. 2007 Jun 29;129(7):1261-74. Review.
4.The E3 ligase TRAF6 regulates Akt ubiquitination and activation.
Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK.
Science. 2009 Aug 28;325(5944):1134-8.
5.Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway.
Morgan TM, Koreckij TD, Corey E.
Curr Cancer Drug Targets. 2009 Mar;9(2):237-49.
6.Modelling the role of the Hsp70/Hsp90 system in the maintenance of protein homeostasis.
Proctor CJ, Lorimer IA.
PLoS One. 2011;6(7):e22038. Epub 2011 Jul 14.
7.The endogenous inhibitor of Akt, CTMP, is critical to ischemia-induced neuronal death.
Miyawaki T, Ofengeim D, Noh KM, Latuszek-Barrantes A, Hemmings BA, Follenzi A, Zukin RS.
Nat Neurosci. 2009 May;12(5):618-26. Epub 2009 Apr 6.
8.Critical role of bad phosphorylation by Akt in cytostatic resistance of human bladder cancer cells.
Szanto A, Bognar Z, Szigeti A, Szabo A, Farkas L, Gallyas F Jr.
Anticancer Res. 2009 Jan;29(1):159-64.
9.Differential involvement of IkappaB kinases alpha and beta in cytokine- and insulin-induced mammalian target of rapamycin activation determined by Akt.
Dan HC, Baldwin AS.
J Immunol. 2008 Jun 1;180(11):7582-9.
10.response element-binding (CREB) protein in Jurkat T leukemia cells treated with TRAIL.
Caravatta L, Sancilio S, di Giacomo V, Rana R, Cataldi A, Di Pietro R.
J Cell Physiol. 2008 Jan;214(1):192-200.
11.p27Kip1 is inactivated in human colorectal cancer by cytoplasmic localization associated with activation of Akt/PKB.
Bottini C, Platini F, Rinaldi M, Leutner M, Alabiso O, Garavoglia M, Tessitore L.
Int J Oncol. 2009 Jan;34(1):69-77.
12.Serum-nutrient starvation induces cell death mediated by Bax and Puma that is counteracted by p21 and unmasked by Bcl-x(L) inhibition.
Braun F, Bertin-Ciftci J, Gallouet AS, Millour J, Juin P.
PLoS One. 2011;6(8):e23577. Epub 2011 Aug 24.
13.XIAP gene expression and function is regulated by autocrine and paracrine TGF-beta signaling.
Van Themsche C, Chaudhry P, Leblanc V, Parent S, Asselin E.
Mol Cancer. 2010 Aug 16;9:216.
14.The WNKs: atypical protein kinases with pleiotropic actions.
McCormick JA, Ellison DH.
Physiol Rev. 2011 Jan;91(1):177-219.
15.Protein kinases and phosphatases in the control of cell fate.
Bononi A, Agnoletto C, De Marchi E, Marchi S, Patergnani S, Bonora M, Giorgi C, Missiroli S, Poletti F, Rimessi A, Pinton P.
Enzyme Res. 2011;2011:329098. Epub 2011 Sep 4.
16.Akt isoforms differentially regulate neutrophil functions.
Chen J, Tang H, Hay N, Xu J, Ye RD.
Blood. 2010 May 27;115(21):4237-46. Epub 2010 Mar 23.
17.PTEN in DNA damage repair.
Ming M, He YY.
Cancer Lett. 2012 Jan 18. [Epub ahead of print]
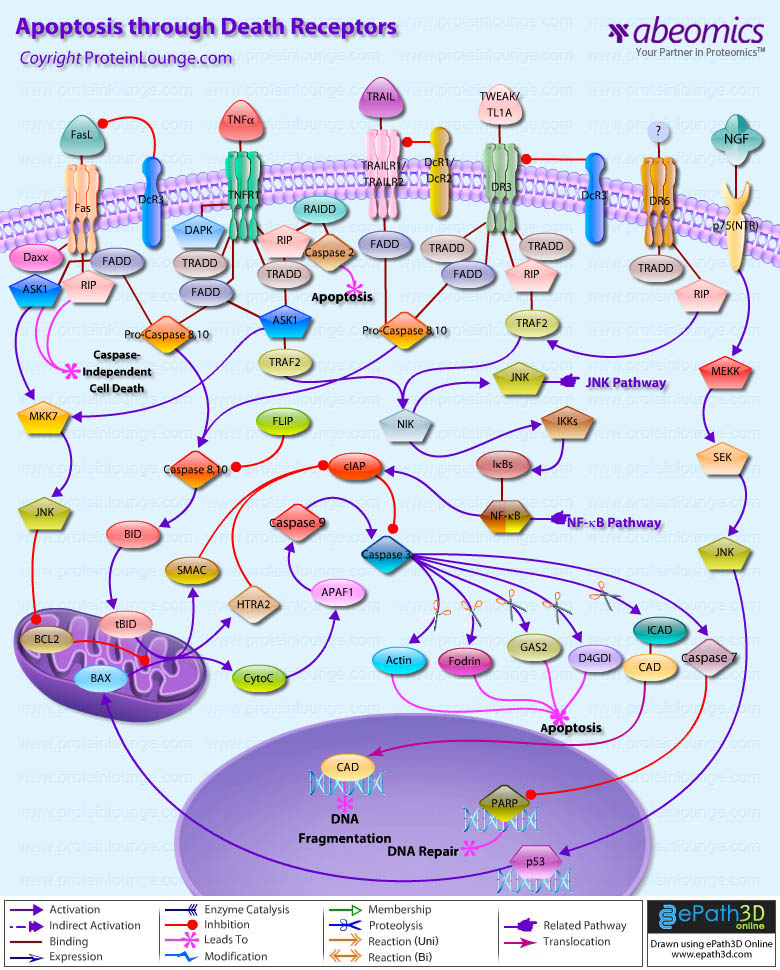
Apoptosis is a cell suicide mechanism that enables metazoans to control cell number in tissues and to eliminate individual cells that threaten the animal’s survival. Certain cells have unique sensors, termed DR (Death Receptors), on their surface, which detect the presence of extracellular death signals and, in response, they rapidly ignite the cell’s intrinsic apoptosis machinery. Death Receptors belong to the TNF (Tumour Necrosis Factor) gene superfamily and generally can have several functions other than initiating apoptosis. Eight members of the Death Receptor family have been characterized so far: TNFR1 (Tumor Necrosis Factor Receptor-1) also known as DR1, CD120a, p55 and p60, Fas (also known as DR2, APO1 and CD95), DR3 (Death Receptor-3) (also known as APO-3, LARD, TRAMP and WSL1), TRAILR1 (TNF-Related Apoptosis-Inducing Ligand Receptor-1) also known as DR4 and APO-2, TRAILR2 (also known as DR5, KILLER and TRICK2), DR6, EDAR (Ectodysplasin-A Receptor) and NGFR (Nerve Growth Factor Receptor). These are distinguished by a cytoplasmic region of approximately 80 residues termed the DD (Death Domain). When these receptors are triggered by corresponding ligands, a number of molecules are recruited to the DD and subsequently a signaling cascade is activated. Death ligands also interact with DcRs (Decoy Receptors) that do not possess DDs and so cannot form signaling complexes. Decoy receptors are members of the TNFR superfamily that are capable of competing with signalling receptors for ligand binding, thereby inhibiting their function. TRAILR3 (also known as DcR1) and TRAILR4 (also known as DcR2) compete with DR4 and DR5 for binding of APO2L/TRAIL. DcR3 competes with Fas for binding of FasL and with DR3 for binding of TL1A. Two types of DR signaling complex can be distinguished. The first group comprises the DISCs (Death-Inducing Signaling Complexes) that are formed at the Fas receptor, TRAILR1 or TRAILR2. All three receptors recruit DISCs with similar compositions. DISC formation results in the activation of Caspase8, which plays the central role in transduction of the apoptotic signal. The second group comprises the TNFR1, DR3, DR6 and EDAR. These recruit a different set of molecules, which transduce both apoptotic and survival signals (Ref.1 & 2).
Fas, a member of the TNFR family, typify the classical view of DR function. The Fas Receptor upon binding to the FasL trimerizes and induces apoptosis through a cytoplasmic domain called DD (Death Domain) that interacts with signaling adaptors like FAF-1 (Fas-Associated Factor-1), FADD (Fas-Associated Death Domain), Daxx, FAP1 (Fas-Associated Protein-Tyrosine Phosphatase-1), Flash and RIP (Receptor-Interacting Protein). FADD carries a DED (Death Effector Domain) and by homologous interaction it recruits the DED containing Procaspase8 protein which is in inactive state. Procaspase8 is proteolytically activated to Caspase8. FLIP (FLICE-Inhibitory Protein) inhibit activation of Procaspase8 at the DISC by blocking its processing. FLIPL also facilitates the cleavage of Procaspase8 at the DISC by forming FLIPL-Procaspase8 heterodimers. FADD also helps in the activation of Caspase10. Caspase8 can cleave the BH3-only protein BID (BH3 Interacting Domain Death Agonist), and the resulting tBID (Truncated BID) can inactivate Bcl2 (B-Cell CLL/Lymphoma-2) in the mitochondrial membrane. This allows the escape of CytoC (Cytochrome-C), which clusters with APAF1 (Apoptotic Protease Activating Factor-1) and Caspase9 in the presence of dATP to activate Caspase9. SMAC (Second Mitochondria-Derived Activator of Caspase)/DIABLO and HTRA2 (High Temperature Requirement Protein-A2) are also released from the mitochondria and inactivates IAPs (Inhibitors of Apoptosis), which further inhibits Caspase3. Active Caspase9 can cleave and activate Procaspase3 to its active form, leading to breakdown of several cytoskeletal and nuclear proteins (structural, signaling proteins or kinases) like GDID4 (GDP-Dissociation Inhibitor-D4), PARP (Poly ADP-Ribose Polymerase), Alpha-Fodrin, GAS2 (Growth Arrest Specific-2) and Lamin-A, thus inducing apoptosis, and degradation of the ICAD (Inhibitor of Caspase-Activated DNase). Besides Fas, TRAILR1and TRAILR2 also leads to apoptosis by formation of DISCs. TRAILR1 and TRAILR2 are activated by binding to the ligand TRAIL. DISCs also consist of oligomerized, probably trimerized, receptors, the DD-containing adaptor molecule FADD, two isoforms of Procaspase8 (Procaspase8/a and Procaspase8/b), Procaspase10 and the cellular FLIPL/S. Signaling downstream of TRAILR1/R2 receptors is similar to Fas signaling (Ref.3 & 4).
TNFR1 signaling differs from Fas Receptor or TRAILR1/R2-induced apoptosis. TNFR1 is an integral membrane protein with its receptor domain exposed at the surface of the cell binding of the complementary death activator, which transmits a signal to the cytoplasm that leads to activation of downstream products. TNFR1, after binding to its ligand TNF-Alpha, recruits TRADD (TNFR-Associated Death Domain) as a platform adaptor, and, in turn, assembles alternative signaling complexes through secondary adaptors. One type of complex is a DISC that involves FADD and Caspase8 (and probably Caspase10) and triggers apoptosis in a manner similar to the other DR. Another complex involves RIP, TRADD, TRAF1/2 (TNFR-Associated Factor), and probably other, as-yet-unidentified molecules. It is proposed to trigger the NF-KappaB signaling pathway through recruitment of the IKK (I-KappaB Kinase) complex and activates JNK1 (c-Jun Kinase) through a TRAF2 and NIK (NF-KappaB-Inducing Kinase) dependent mechanism. ASK1 (Apoptosis signal-regulating Kinase-1), a MAP Kinase Kinase Kinase, is also required for TNF-mediated JNK activation. TNFR1 is also able to mediate apoptosis through the recruitment of an adapter molecule called RAIDD (RIP-associated ICH-1 / CED-3 homologous protein with a death domain). RAIDD associates with RIP through interactions between death domains and can recruit Caspase2 through an interaction with a motif, similar to the death effector domain, known as CARD (caspase recruitment domain). Recruitment of Caspase2 leads to induction of apoptosis. Another kinase, which is believed to be involved in TNF-Alpha induced apoptosis is DAPK (Death-Associated Protein Kinase). DAPK is a Calcium/Calmodulin regulated serine/threonine Kinase that carries ankyrin repeats, a death domain, and is localized to the cytoskeleton (Ref.5 & 6).
The DR3 and DR6 signaling pathways are less well characterized. Ligand for DR3 is TWEAK (TNF-Related Weak Inducer of Apoptosis)/TL1A while the ligand for DR6 remains undiscovered. RIP and TRADD are recruited to the receptor complex, and DR3 and DR6 promote activation of NF-KappaB that leads to the expression of survival genes. Another death receptor, which plays an important role in nervous system is p75(NTR). p75(NTR) is a member of the TNFR (Tumor Necrosis Factor) receptor superfamily, having no tyrosine kinase domain. NGF binding selectively to p75(NTR) brings about the activation of the proapoptotic JNK cascade. MEKKs (MAP/ERK Kinase Kinases) and SEK (SAPK/ERK Kinase) are the upstream regulators of JNK. JNKs, in turn, upregulate p53 and the proapoptotic member of the Bcl2 family: BAX (Bcl2 Associated-X Protein)in a sequential manner, which bring about apoptosis of the neuronal cells. Our current understanding of DR signaling has opened possibilities for the design of new therapeutic strategies for targeting death receptor pathways. This would allow the treatment of a number of diseases potentially associated with defects in DR signaling, such as multiple sclerosis and Alzheimer’s disease. In tumours that retain some responsiveness to conventional therapy, death-receptor engagement in combination with chemotherapy or irradiation might lead to synergistic apoptosis activation, and reduce the probability that tumour cells that are resistant to either type of agent will emerge. In tumours that have lost p53 function, death-receptor targeting might help to circumvent resistance to chemotherapy and radiotherapy (Ref.7, 8 & 9).
References:
1.Fas death receptor signalling: roles of Bid and XIAP.
Kaufmann T, Strasser A, Jost PJ.
Cell Death Differ. 2012 Jan;19(1):42-50. doi: 10.1038/cdd.2011.121. Epub 2011 Sep 30.
2.Death receptor signal transducers: nodes of coordination in immune signaling networks.
Wilson NS, Dixit V, Ashkenazi A.
Nat Immunol. 2009 Apr;10(4):348-55. Epub 2009 Mar 19.
3.Caspases – an update.
Chowdhury I, Tharakan B, Bhat GK.
Comp Biochem Physiol B Biochem Mol Biol. 2008 Sep;151(1):10-27. Epub 2008 Jul 3.
4.Regulating TRAIL receptor-induced cell death at the membrane : a deadly discussion.
Shirley S, Morizot A, Micheau O.
Recent Pat Anticancer Drug Discov. 2011 Sep;6(3):311-23.
5.Regulation of death receptor signaling by the ubiquitin system.
Wertz IE, Dixit VM.
Cell Death Differ. 2010 Jan;17(1):14-24.
6.Caspase-2: the orphan caspase.
Bouchier-Hayes L, Green DR.
Cell Death Differ. 2012 Jan;19(1):51-7. doi: 10.1038/cdd.2011.157. Epub 2011 Nov 11.
7.The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases.
Meylan F, Davidson TS, Kahle E, Kinder M, Acharya K, Jankovic D, Bundoc V, Hodges M, Shevach EM, Keane-Myers A, Wang EC, Siegel RM.
Immunity. 2008 Jul 18;29(1):79-89. Epub 2008 Jun 19.
8.TL1A and DR3, a TNF family ligand-receptor pair that promotes lymphocyte costimulation, mucosal hyperplasia, and autoimmune inflammation.
Meylan F, Richard AC, Siegel RM.
Immunol Rev. 2011 Nov;244(1):188-96. doi: 10.1111/j.1600-065X.2011.01068.x.
9.p53 modulates acquired resistance to EGFR inhibitors and radiation.
Huang S, Benavente S, Armstrong EA, Li C, Wheeler DL, Harari PM.
Cancer Res. 2011 Nov 15;71(22):7071-9. Epub 2011 Nov 8.
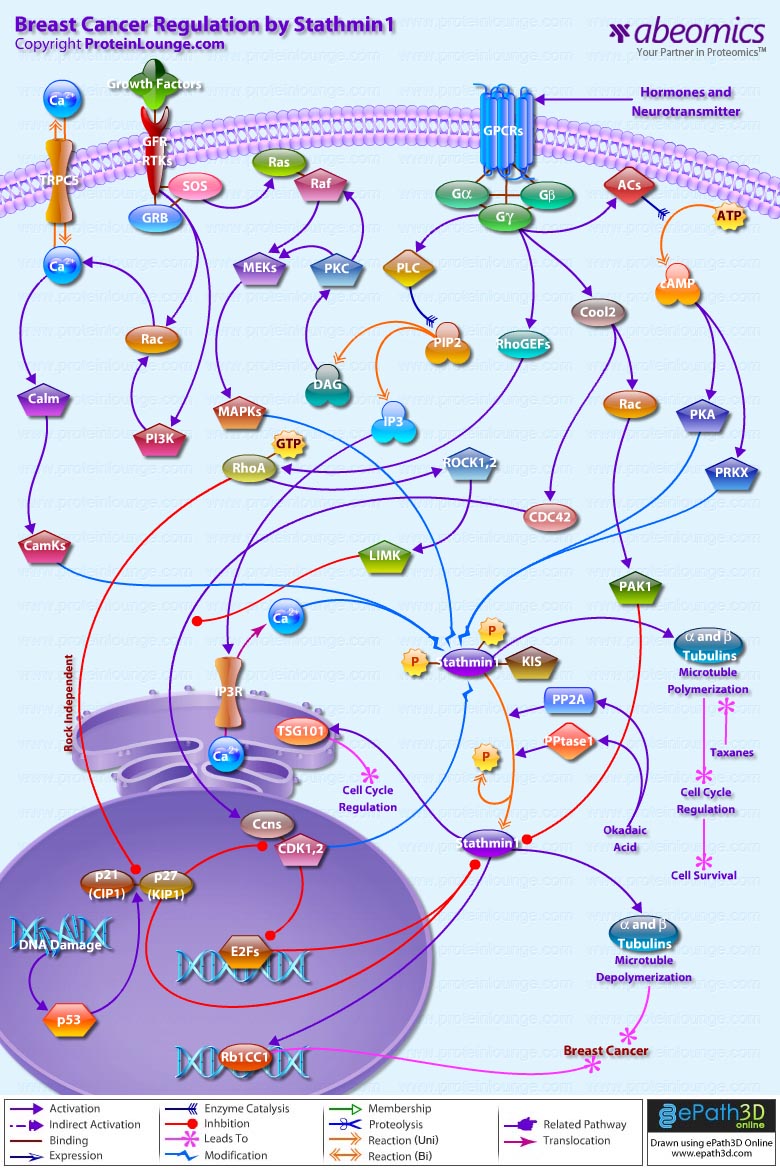
Stmn1 (Stathmin-1) also referred to as Op18 (Oncoprotein-18) is a major regulator of microtubule dynamics. It is an evolutionarily well conserved 17 kDa cytoplasmic phosphoprotein that is highly expressed in a wide variety of cancers and its high abundance seems to be necessary for the maintenance of the transformed phenotypes. Breast cancers exhibit high levels of Stmn1 and may be resistant to anti-microtubule agents. Stmn1 destabilizes microtubule polymers of Alpha and Beta-Tubulin subunits, by promoting catastrophes that ultimately results in deregulation of cell cycle, hampering cell survival (Ref.1). One of the key properties of microtubules is that of ‘dynamic instability’. Dynamic instability comprises the continuous switching between catastrophes (depolymerization or shrinkage phase) and rescues (polymerization or growing phase) of individual microtubules. Stmn1 expression is ubiquitous and is phosphorylated on up to four sites (Ser16 (Serine-16), Ser25, Ser38 and Ser63) in response to many regulatory signals within cells, in order to maintain equilibrium during microtubule polymerization. Its molecular characterization indicates a functional organization including an N-terminal regulatory domain that bears the phosphorylation sites, linked to a putative Alpha-helical binding domain predicted to participate in coiled-coil, protein-protein interactions (Ref.2).
In addtion to the protein kinases that phosphorylate Stmn1 such as CamKs (Calcium/Calmodulin-Dependent Protein Kinases), MAPKs (Mitogen-Activated Protein Kinases), CDKs (Cyclin-Dependent Kinases), PKA (cAMP dependent Protein Kinase-A), PRKX (Protein Kinase-X-Linked); a few other proteins are known to interact with Stmn1 in vivo. One of them is a putative Serine/Threonine kinase, KIS (Kinase Interacting Stathmin), which binds to Stmn1 and is regulated by it or, more likely, is a part of the kinases controlling phosphorylation state of Stmn1. Two other proteins, Rb1CC1 (Rb1-Inducible Coiled-Coil-1) and TSG101 (Tumor Susceptibility Gene-101), form Alpha-helices participating in coiled-coil interacting structures to regulate cell cycle (Ref.3 & 4). The upstream factors that activate the kinases modulating Stmn1 function, includes activation by hormones and neurotransmitters through GPCRs (G-Protein Coupled Receptors); activation by Growth Factors through Growth Factor Receptors and RTKs (Receptor Tyrosine Kinases); and Ca2+ (Calcium) influx through channel proteins like TRPC5 (Transient Receptor Potential Cation Channel Subfamily-C Member-5) and IP3Rs (IP3 Receptors). Hormones and neurotransmitters stimulate the G-proteins (Alpha, Beta and Gamma-subunits) that activate downstream effectors like ACs (Adenylate Cyclases), Cool2, RhoGEFs (Rho Guanine Nucleotide Exchange Factors) and PLC (Phospholipase-C) in order to intensify phoshorylation of Stmn1. The tmACs (Trans-membrane Adenylate Cyclase) enzymes are responsible for the accumulation of cAMP (Cyclic Adenosine 3′,5′-monophosphate) from ATP (Adenosine Triphosphate). cAMP directly regulates the activities of PKA (cAMP-dependent Protein Kinase-A) and PRKX (Protein Kinase-X-Linked) leading to phosphorylation of Stmn1 at Ser16 and Ser63. G-proteins activate PLC (Phospholipase-C) to cleave PIP2 (Phosphatidylinositol 4,5-bisphosphate) generating DAG (Diacylglycerol) and IP3 (Inositol 1,4,5-trisphosphate). DAG activates PKC (Protein Kinase-C), whereas, IP3 modulates Ca2+ release through IP3R (IP3 Receptor). Activation of PKC acts in concert with Growth Factor Receptor/RTK associated proteins like GRB (Growth Factor Receptor-Bound Proteins) and SOS (Son of Sevenless) to phosphorylate Stmn1 at Serine position 25 through a signaling cascade that includes, Ras, Raf, MEKs (MAPK/ERK Kinases), and MAPKs (Mitogen-Activated Protein Kinases). GRB and SOS also activate Rac and PI3K (Phosphatidylinositde-3-Kinase) to modulate release of Ca2+ ions through TRPC5 (Transient Receptor Potential Cation Channel Subfamily-C Member-5). This further activates Calm (Calmodulin) and CamKs (Calcium/Calmodulin-Dependent Protein Kinases), which along with Ca2+ ions act synergistically with PKA and PRKX to phosphorylate Stmn1 at Ser16 (Ref.5 & 6).
Further G-protein signaling couples Rac and CDC42 (Cell Division Cycle-42) activation, with RhoA activation, to phosphorylate and inactivate Stmn1. G-proteins stimulate Guanine Nucleotide Exchange Factors like RhoGEFs and Cool2 to activate RhoA and Rac-CDC42, respectively. Rac and CDC42 relay signals to PAK1 (p21/CDC42/Rac1-Activated Kinase-1) and Ccns (Cyclins) that in turn inhibit anti-microtubule action of Stmn1 through phosphorylations (Ref.7). Rac-CDC42 and RhoA function in parallel pathways to stimulate Ccn expression. The targets of RhoA include Rho-Kinase/ROCK (Rho-Associated Coiled-Coil-Containing Protein Kinase) and LIMK (LIM Kinase). Activity of Rho-ROCK-LIMK signaling promotes Ccn expression and suppresses Rac-CDC42 activation of Ccn. So, reducing Rho-ROCK-LIMK signaling would stimulate Rac-CDC42 dependent Ccn expression (Ref.3).
RhoA acting independently of ROCK, also modulates the cell cycle by regulating expression of p21(CIP1) (Cyclin Dependent Kinase Inhibitor-p21) and p27(KIP1) (Cyclin Dependent Kinase Inhibitor-p27) and this reflects on Stmn1 phosphorylation and Tubulin dynamics. p27(KIP1) binds to RhoA and prevents the interaction of RhoA with its activators. The transcriptional regulation of p21(CIP1) and p27(KIP1) is controlled by p53 and this signifies the up-regulation of Stmn1 in human breast cancers, under conditions like mutations or DNA damage (Ref.8). p27(KIP1) has the potential for modulating Stmn1 expression and/or function through at least four different mechanisms: (i) p27(KIP1) might directly interact with Stmn1 and prevent its microtubule destabilizing activity; (ii) p27(KIP1) may activate Rac, which can promote the phosphorylation of Stmn1 and inhibit its microtubule destabilizing activity; (iii) the inhibitory effect of p27(KIP1) on CDKs might result in either E2F transcription factor mediated down regulation of Stmn1 expression or increased Stmn1 activity by means of CDK inactivation; (iv) p27(KIP1) and Stmn1 might interact through KIS that phosphorylates p27(KIP1) and induces its translocation to the cytoplasm where Stmn1 is localized (Ref.9 & 10). However dephosphorylation of Stmn1 by an Okadaic Acid-sensitive PPtase1 (Protein Phosphatase-1) and PP2A (Protein Phosphatase-2A) is crucial for the ability of cells to exit mitosis. A similar effect is also shown by Taxanes, group of chemotherapy drugs that includes Taxol and Taxotere, specifically used to kill cancer cells of mammary gland by stopping their growth (Ref.11).
Stmn1 is a major microtubule-destabilizing protein that promotes microtubule depolymerization by two distinct mechanisms. The first is a catastrophe-promoting microtubule depolymerization activity that is necessary for the regulation of the mitotic spindle. The second is a Tubulin-sequestering activity that inhibits microtubule polymerization and is necessary for the regulation of microtubule dynamics during interphase (Ref.12). Both activities are regulated by phosphorylation, which inactivates Stmn1 and prevents its binding to Tubulin and interferes with the sensitivity of breast cancer cells to anti-microtubule drugs (Ref.13).
References:
1.The oncoprotein 18/stathmin family of microtubule destabilizers.
Cassimeris L.
Curr. Opin. Cell Biol. 2002 Feb;14(1):18-24.
2.The effect of stathmin phosphorylation on microtubule assembly depends on tubulin critical concentration.
Amayed P, Pantaloni D, Carlier MF.
J. Biol. Chem. 2002 Jun 21;277(25):22718-24.
3.Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors?
Besson A, Assoian RK, Roberts JM.
Nat. Rev. Cancer. 2004 Dec;4(12):948-55.
4.Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas.
Liu RT, Huang CC, You HL, Chou FF, Hu CC, Chao FP, Chen CM, Cheng JT.
Oncogene. 2002 Jul 18;21(31):4830-7.
5.Identification of Op18/stathmin as a potential target of ASK1-p38 MAP kinase cascade.
Mizumura K, Takeda K, Hashimoto S, Horie T, Ichijo H.
J. Cell Physiol. 2005 Aug 18.
6.The PI 3-kinase-Rac-p38 MAP kinase pathway is involved in the formation of signet-ring cell carcinoma.
Xu Q, Karouji Y, Kobayashi M, Ihara S, Konishi H, Fukui Y.
Oncogene. 2003 Aug 28;22(36):5537-44.
7.Conserved microtubule-actin interactions in cell movement and morphogenesis.
Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM.
Nat. Cell Biol. 2003 Jul;5(7):599-609.
8.Overexpression of stathmin in breast carcinomas points out to highly proliferative tumours.
Curmi PA, Nogues C, Lachkar S, Carelle N, Gonthier MP, Sobel A, Lidereau R, Bieche I.
Br. J. Cancer. 2000 Jan;82(1):142-50.
9.p27(Kip1) and stathmin share the stage for the first time.
Iancu-Rubin C, Atweh GF.
Trends Cell Biol. 2005 Jul;15(7):346-8.
10.E2F mediates sustained G2 arrest and down-regulation of Stathmin and AIM-1 expression in response to genotoxic stress.
Polager S, Ginsberg D.
J. Biol. Chem. 2003 Jan 17;278(3):1443-9.
11.Stathmin inhibition enhances okadaic acid-induced mitotic arrest: a potential role for stathmin in mitotic exit.
Mistry SJ, Atweh GF.
J. Biol. Chem. 2001 Aug 17;276(33):31209-15.
12.The role of stathmin in the regulation of the cell cycle.
Rubin CI, Atweh GF.
J. Cell Biochem. 2004 Oct 1;93(2):242-50.
13.Effect of stathmin on the sensitivity to antimicrotubule drugs in human breast cancer.
Alli E, Bash-Babula J, Yang JM, Hait WN.
Cancer Res. 2002 Dec 1;62(23):6864-9.
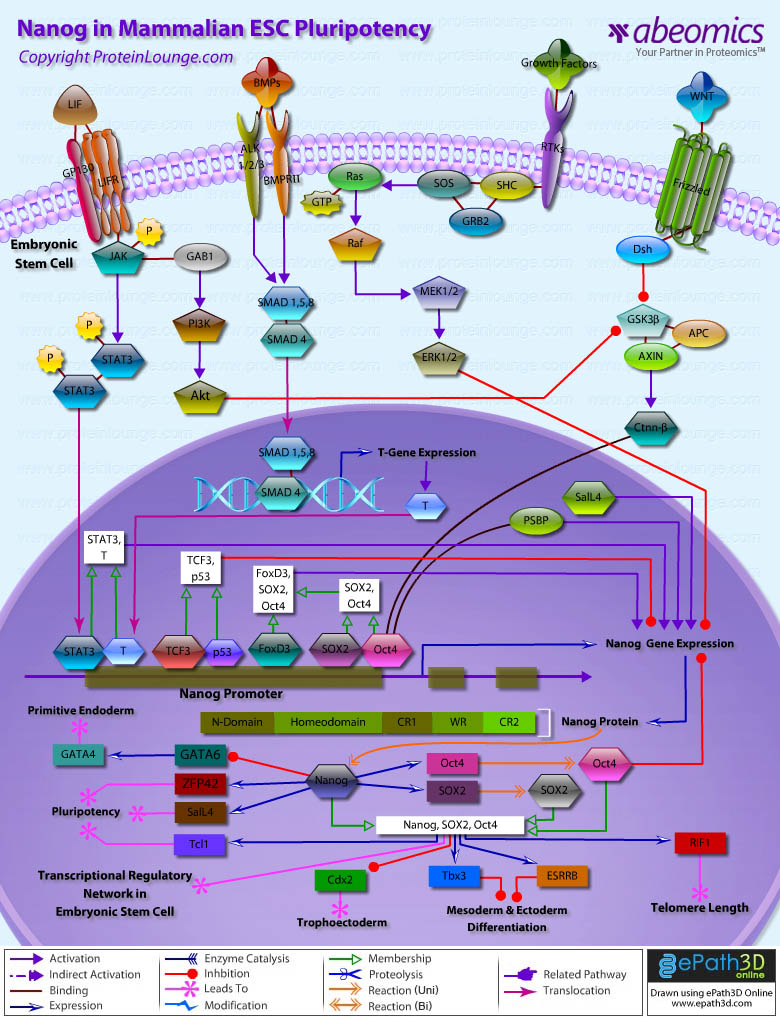
ESCs (Embryonic stem cells) are Pluripotent cells derived from the ICM (Inner Cell Mass) of Blastocyst-stage embryos. These cells have two distinctive properties: an unlimited capacity for Self-renewal and Pluripotency. The capability for Self-renewal and the Pluripotency of ESCs seem to be under the control of multiple transcriptional factors, most common among them being Nanog (Nanog homeobox), Oct4 (Octamer Binding Transcription Factor-4) and SOX2 (SRY (Sex Determining Region-Y) Box-2). Functions of these transcription factors depend on the stage of development of a Pluripotent cell, indicating that these factors function in combination with other processes. The activity of these transcription factors also depends on the accessibility of their target genes, which are made accessible by the modification of their DNA, histones, or chromatin structure. These transcription factors activate or repress patterns of gene expression that mediate phenotypic changes during Stem Cell differentiation (Ref.1 & 2).
The transcription factor Nanog is a homeodomain-bearing protein that is transcribed specifically in Pluripotent cells in Mouse preimplantation embryos, ESCs, and EGCs (Embryonic Germ Cells) and Monkey and Human ESCs. Nanog protein consists of three domains with the homeodomain separating the N-terminal serine-rich 96 amino acids from the C-terminal 150 amino acids. Both N- and C-terminal domains in Mouse Nanog show trans-activator function, but the activity of the C-terminal domain is much higher than the N-terminal domain. In Human Nanog, transactivation potential is retained in the C-terminal domain but not in the N-terminal domain. The prominent feature of the C-terminal domain is the presence of a 10 pentapeptide repeat (WR), each starting with a Tryptophan (W). This repeat is well conserved between Human and Mouse, although one of the tryptophans is replaced by a Glutamine (Q) in Human Nanog. In addition to WR, the C-terminal domain also contains another trans-activator module, the 58 amino acids region C-terminal to the WR (CD2). Both WR and CD2 can separately transactivate the expression of a reporter gene (Ref.3 & 4).
Nanog expression is restricted to Pluripotent cells and is downregulated upon differentiation, but little is known about how Nanog expression is regulated. Nanog gene is transcribed under the control of a regulatory region that lies within 332 bp upstream of the transcriptional start site. This region contains Octamer and SOX elements, which are highly conserved among the 5’-flanking regions of the Mouse, Monkey, and Human Nanog genes. Oct4 and SOX2 are the major transcription factors that bind to the Nanog promoter in vitro and in vivo to promote Nanog transcription. However, high levels of Nanog are beneficial to ES cell self-renewal, but overexpressed Oct4 induces differentiation. Other Pluripotent factors also contribute to the regulation of Nanog expression. FoxD3 (Forkhead Box Protein-D3), a Forkhead family transcription factor, is highly expressed in Mouse ES cells and in Pluripotent cells of the early embryo. FoxD3 activates the Nanog promoter through an ESC-specific enhancer localized at –270 upstream of the transcription start site. Nanog is also a direct downstream effector of the LIF (Leukaemia Inhibitory Factor)-STAT3 (Signal Transducer and Activator of Transcription-3) pathway in maintaining ESC Pluripotency. Besides, BMP (Bone Morphogenetic Protein) Pathway also regulates Nanog expression. In the absence of LIF, low concentrations of BMPs promote mesoderm differentiation of Mouse ESCs with upregulation of the mesoderm marker-T (Brachyury) and prevent the neural-ectoderm differentiation. In the presence of LIF, STAT3 activated by JAK (Janus tyrosine Kinase), which is recruited by LIFR–GP130 (GP130 Transducer Chain) heterodimer on binding LIF, interacts with Brachyury and binds the Nanog promoter, resulting in upregulation of Nanog expression. BMPs belong to the TGF-Beta (Transforming Growth Factor-Beta) Superfamily and mediate signaling through their downstream effectors – SMAD1 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-1) or the closely related SMAD5 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-5) and SMAD8 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-8). Nanog can physically interact with SMAD1 and interfere with the further recruitment of the coactivators to the active SMAD1 complexes, thus inhibiting the activity of BMP signaling and limiting mesoderm progression and ultimately maintaining the undifferentiated state of Mouse ESCs (Ref.5 & 6). Ctnn-Beta (Catenin-Beta), a key regulator of WNT (Wingless-Type MMTV Integration Site Family Member) Pathway, also upregulates Nanog. In general, Ctnn-Beta stability is negatively regulated through its phosphorylation by GSK3Beta (Glycogen Synthase Kinase-3-Beta), APC (Adenomatous Polyposis Coli) and AXIN (Axis Inhibitor) complex. Extracellular WNT proteins activate the canonical WNT signaling pathway by binding the Fz (Frizzled) seven transmembrane span receptor and stabilizing intracellular Ctnn-Beta, which then translocates to the nucleus where it interacts with TCF (T-Cell Factor)/ LEF (Lymphocyte Enhancer Factor) transcription factors to regulate expression of target genes. Ctnn-Beta can also be stabilized by LIF Pathway. LIF stimulation leads to inactivation of GSK3Beta through the PI3K (Phosphoinositide 3-Kinase)/Akt pathway, thus stabilizing Ctnn-Beta. Ctnn-Beta is involved in upregulation of Nanog through binding with Oct3/4 protein in nucleus. Oct4 can also bind to a novel PSBP (Pluripotential cell-specific SOX element-Binding Protein) leading to Nanog gene expression. Oct4 dominantly bound to the Octamer element, while PSBP preferentially bound to the SOX element. SalL4 (Sal-Like-4 (Drosophila)), a member of Spalt-like protein family, interacts with Nanog and exists as a complex with Nanog. SalL4 and Nanog complex resembles the network configuration for the Oct4 and SOX2 complex. Nanog and SalL4 co-targets many genomic sites. SalL4 and Nanog also bind to and regulate the respective regulatory regions of their own genes (Ref.1, 7, 8 & 9).
Once expressed, Nanog blocks differentiation. Thus, negative regulation of Nanog is required to promote differentiation during embryonic development. The tumor suppressor protein p53 binds to the Nanog promoter and act as negative regulator of Nanog during ESC differentiation. TCF3 (Transcription Factor-3), a transcription factor that functions downstream of the WNT pathway bind to a regulatory region on the Nanog promoter and repress the promoter activity in ESCs. Several signaling molecules are also involved in Nanog repression by primitive endoderm specification. Major signaling pathway involved in primitive endoderm differentiation is the GRB2 (Growth Factor Receptor-Bound Protein-2)/SOS (Son of Sevenless)/Ras/ERK (Extracellular Signal-Regulated Kinase) pathway. The canonical ERK cascade is stimulated upon the binding of extracellular Growth factors to their respective transmembrane RTKs (Receptor Tyrosine Kinases). The subsequent auto-phosphorylation of the cytoplasmic tails of the receptor on tyrosine leads to the tyrosine phosphorylation of the adapter protein SHC. SHC can then recruit the GRB2-SOS complex to the membrane via the SH2 domain of GRB2 binding to the phosphotyrosine on SHC. SOS, a GEF for Ras, can then exchange the GDP bound to Ras to GTP. Once Ras binds GTP, it can then recruit the serine/threonine kinase Raf to the membrane. When Raf translocate to the membrane, it becomes activated and then phosphorylates the dual specificity kinases MEK1 (MAPK/ERK Kinase-1) and MEK2 (MAPK/ERK Kinase-1). MEKs (MAPK/ERK Kinases) then phosphorylate ERK1/2, which then inhibit Nanog expression (Ref.10, 11 & 12).
The mechanism through which Nanog regulates Stem cell Pluripotency is not known properly. Nanog may assume a bifunctional role to repress those genes important for differentiation and activate the ones necessary for self-renewal. Nanog can regulate the expression of REX1/ZFP42 (Zinc Finger Protein-42) by binding to its promoter. This regulation is likely mediated by the C-terminal transactivator of Nanog. SOX2, not Oct3/4, cooperate with Nanog in regulating REX1 activity. GATA4 (GATA Binding Protein-4) and GATA6 (GATA Binding Protein-6), endoderm transcription factors are also regulated by Nanog. Nanog directly repress GATA6, which results in repression of GATA4, thereby inhibiting ESC differentiation. Nanog also regulate the expression of Oct4 and SOX2 genes which play important role in Pluripotency. Nanog also activate functions in concert with other factors such as Oct4 and SOX2 to establish ESC identity. These factors regulate a core transcriptional circuit for ESC self-renewal. The major downstream target in Mouse includes ESRRB (Estrogen Related Receptor-Beta) and RIF1 (Rap1 Interacting Factor-1 homolog (yeast)). ESRRB belongs to the superfamily of nuclear hormone receptors, and is necessary to block the differentiation into mesoderm, ectoderm and neural crest cells but are not required to repress trophectoderm differentiation. RIF1 is an ortholog of a yeast telomeric protein and is upregulated in Mouse ES and germ cells. Other genes regulated by Nanog include Tbx3 (T-box 3) and TCL1 (T-Cell Lymphoma breakpoint-1). The function of TCL1 is to repress only a subset of neural crest genes, whereas Tbx3 blocks the mesoderm and ectoderm differentiation. Downregulation of Nanog, SOX2, ESRRB, Tbx3 or TCL1 leads to the immediate induction of Otx2 (Orthodenticle homolog 2 (Drosophila)), Pitx2 (Paired-like homeodomain Transcription factor-2), Sox18 (SRY (Sex determining region Y)-box 18) and probably additional genes. All three genes are expressed in the epiblast. Oct4, SOX2 and Nanog also repress Caudal-type homeodomain transcription factor Cdx2 ((Caudal type homeobox-2)) expression. Cdx2 is necessary for trophectoderm development. In undifferentiated Human ES cells, about 352 genes are bound by Oct4, Nanog and SOX2 simultaneously, which may be expressed or repressed (Ref.13, 14 & 15). The ability of Nanog when overexpressed to robustly maintain ESC identity, without the normally obligatory LIF and BMP/GDF Growth Differentiation Factor) signals, places Nanog firmly at the hub of the circuitry responsible for the properties of Self-renewal and Pluripotency. The mechanisms of Nanog function and regulation within these circuits are at present poorly perceived. However, as genes acting both upstream and downstream of Nanog are uncovered, and protein partners of Nanog are identified, a clearer vision is emerging of how Nanog functions. Identification of Nanog is a crucial step in understanding early embryogenesis and in exploiting Pluripotent cells for therapeutic goals (Ref.1 & 16).
References:
1.Transcriptional dynamics of the embryonic stem cell switch.
Chickarmane V, Troein C, Nuber UA, Sauro HM, Peterson C.
PLoS Comput Biol. 2006 Sep 15;2(9):e123.
2.Core transcriptional regulatory circuitry in human embryonic stem cells.
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA.
Cell. 2005 Sep 23;122(6):947-56.
3.Identification of a putative transactivation domain in human Nanog.
Oh JH, Do HJ, Yang HM, Moon SY, Cha KY, Chung HM, Kim JH.
Exp Mol Med. 2005 Jun 30;37(3):250-4.
4.Nanog and transcriptional networks in embryonic stem cell pluripotency.
Pan G, Thomson JA.
Cell Res. 2007 Jan;17(1):42-9. Review.
5.Transcriptional regulation of nanog by OCT4 and SOX2.
Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P.
J Biol Chem. 2005 Jul 1;280(26):24731-7.
6.Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression.
Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T.
Mol Cell Biol. 2005 Mar;25(6):2475-85.
7.Beta-catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells.
Takao Y, Yokota T, Koide H.
Biochem Biophys Res Commun. 2007 Feb 16;353(3):699-705. Epub 2006 Dec 20.
8.Regulation of Nanog expression by phosphoinositide 3-kinase-dependent signaling in murine embryonic stem cells.
Storm MP, Bone HK, Beck CG, Bourillot PY, Schreiber V, Damiano T, Nelson A, Savatier P, Welham MJ.
J Biol Chem. 2007 Mar 2;282(9):6265-73.
9.Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells.
Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, Ng HH.
J Biol Chem. 2006 Aug 25;281(34):24090-4.
10.Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal.
Pereira L, Yi F, Merrill BJ.
Mol Cell Biol. 2006 Oct;26(20):7479-91.
11.Regulation of apoptosis and differentiation by p53 in human embryonic stem cells.
Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, Wu J, Ding M, Deng H.
J Biol Chem. 2007 Feb 23;282(8):5842-52.
12.The Grb2/Mek pathway represses Nanog in murine embryonic stem cells.
Hamazaki T, Kehoe SM, Nakano T, Terada N.
Mol Cell Biol. 2006 Oct;26(20):7539-49.
13.Pluripotential competence of cells associated with Nanog activity.
Hatano SY, Tada M, Kimura H, Yamaguchi S, Kono T, Nakano T, Suemori H, Nakatsuji N, Tada T.
Mech Dev. 2005 Jan;122(1):67-79.
14.Maintenance of embryonic stem cell pluripotency by Nanog-mediated reversal of mesoderm specification.
Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gage FH, Rodriguez-Esteban C, Belmonte JC.
Nat Clin Pract Cardiovasc Med. 2006 Mar;3 Suppl 1:S114-22.
15.The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells.
Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S.
Cell. 2003 May 30;113(5):631-42.
16.A heterogeneous expression pattern for Nanog in embryonic stem cells.
Singh AM, Hamazaki T, Hankowski KE, Terada N.
Stem Cells. 2007 Oct;25(10):2534-42.
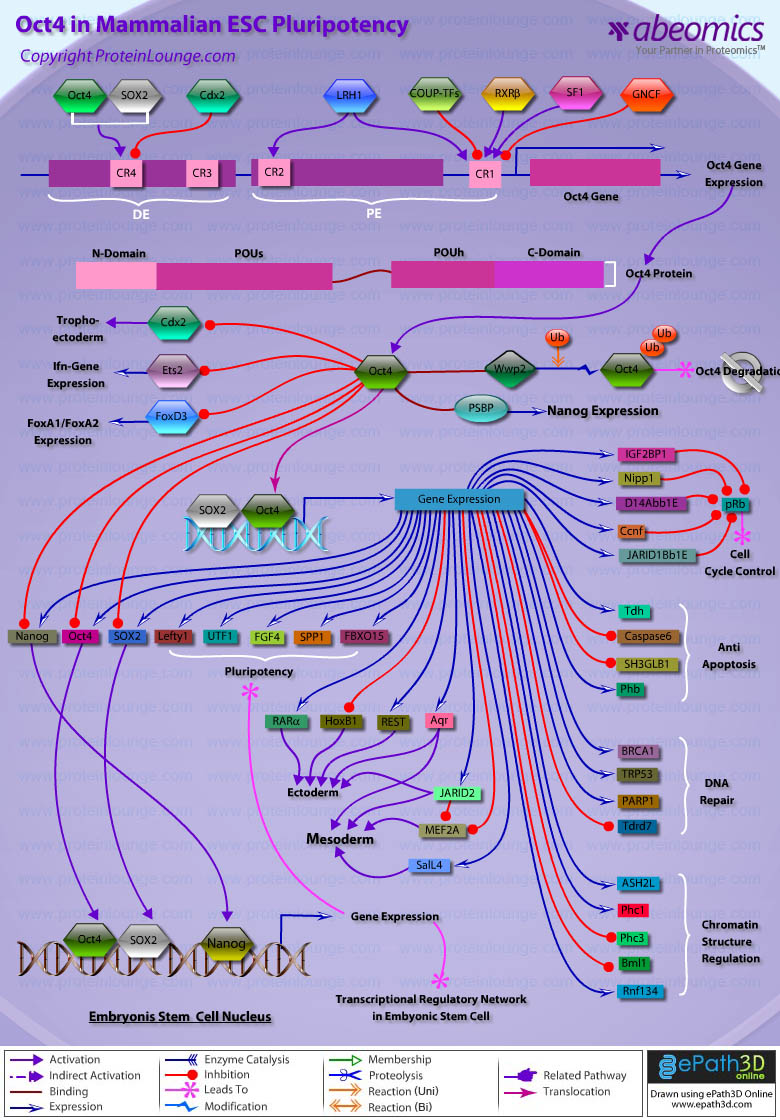
Stem cells are undifferentiated cells capable of producing virtually all cell types in our body. They are characterized by the ability to Self-renew and maintain Pluripotency. For proper developmental outcome, ESCs (Embryonic Stem Cells) must tightly regulate their differentiation status. Hundreds of genes have been identified, including several transcription factors, which have expression patterns tightly correlated with ESC differentiation. Very low number of transcription factors is responsible for regulating the development of an entire organism. These transcription factors form multiprotein complexes on DNA, thereby orchestrating the correct temporal–spatial expression of developmental genes. The process leads to the establishment of functional partnerships, with the combination rather than the individual activity of each factor eliciting specific transcriptional outcomes. Two key transcription factors, Oct4 (Octamer Binding Transcription Factor-4) and Nanog, have been identified that are crucial for maintenance of the Pluripotent state of ESCs (Ref.1 & 2).
Oct4, also known as Oct3, is a homeodomain transcription factor of the POU (Pit-Oct-Unc) family. The POU family of transcription factors can activate the expression of their target genes through binding an octameric sequence motif of an AGTCAAAT consensus sequence. Oct4 (POU5F1) plays a central role in Self-renewal, Pluripotency, and Lineage commitment in ESCs, the Embryonic Epiblast and PGCs (Primordial Germ Cells). Oct4 expression levels should be strictly maintained to regulate cell fate. Alterations in Oct4 expression promote differentiation and leads to the specification of Ectodermal, Endodermal, or Mesodermal primitive progenitors. Oct4 Protein consists of 3 domains: N-terminal domain, POU domain and a C-Terminal domain. POU domain consists of two structurally independent subdomains: a 75 amino acid amino-terminal POU specific (POUs) region and a 60 amino-acid carboxyl-terminal homeodomain (POUh). Both domains make specific contact with DNA through a helix-turn helix structure and are connected by a variable linker of 15 to 56 amino-acids. Regions outside the POU domain are not critical for DNA binding and exhibit little sequence conservation. The N-terminal domain (N-domain) is rich in Proline and Acidic residues, while the C-terminal domain (C-domain) is rich in Proline, Serine and Threonine residues. The N-domain plays an important role in transactivation. The C-domain also plays a role in transactivation. The activity of Oct4 C-domain is cell type specific and is regulated through phosphorylation, whereas the N-domain is not. Oct4 POU-domain functions differently by serving as interaction sites for cell type-specific regulatory factors (Ref.3 & 4).
The precise mechanisms by which the activation and expression of Oct4 genes are regulated still remain to be elucidated. To date, two regulatory elements, a Distal and a Proximal Enhancer, have been identified as stem-cell-specific enhancers of Oct3/4 gene, to which many positive and negative regulators are recruited. There are 4 regions that are highly conserved among Human, Bovine and Mouse Oct4 promoter/enhancer elements, designated as CR1 (Conserved Region-1), CR2, CR3 and CR4 respectively. CR1 is downstream of Proximal Enhancer and immediately upstream of Exon 1. Each enhancer contains multiple potential binding sites for transcription factors that can either activate or repress Oct4 expression. In addition, methylation in these regions represses Oct4 expression in differentiated cells. Several positive and negative regulators bind to the Oct4 gene to regulate its expression. Among them, members of the orphan nuclear receptor superfamily, which can bind to the Proximal Enhancer, are known to influence Oct3/4 expression. LRH-1 (Liver Receptor Homolog-1), also known as NR5A2, is a putative positive regulator of Oct3/4. Other positive regulators include SF1 (Steroidogenic Factor-1) and RXR-Beta (Retinoid X Receptor-Beta). By contrast, GCNF (Germ Cell Nuclear Factor) or NR6A1 is a potential Oct3/4 negative regulator. COUF-TFI/II (Chicken Ovalbumin Upstream promoter-Transcription Factors- I/II), encoded by NR2F1 and NR2F2, respectively, also function as negative regulators of Oct3/4 expression. The balance between these positive and negative regulators might determine the precise level of Oct3/4 expression in response to extracellular stimuli. Oct4 can also be degraded by interacting with a Murine E3 Ub ligaseWwp2 (WW domain containing E3 Ubiquitin Protein ligase-2). Wwp2 can directly bind to Oct4 through WW domains. It promotes ubiquitination of Oct4 both in vitro and in vivo (Ref.1, 5 & 6).
Oct4 protein, once activated can act as an activator or repressor of target genes to maintain stem cell pluripotency. Oct4 usually functions in concert with other regulators to activate specific target genes in specific cell types at defined developmental stages. Most common regulators that cooperate with Oct4 in activating Oct3/4 target genes include SOX2 (SRY (Sex Determining Region-Y) Box-2) and Nanog. SOX2 is an HMG domain-containing transcription factor that binds to the consensus Motif CATTGTT. Oct4 and SOX2 bind to a few thousand regulatory sites in the ESC genome. They reciprocally regulate POU5F1 and SOX2 transcription via the Oct4-SOX2 complex in ESCs. In addition, Oct4 and SOX2 positively regulate Nanog, revealing that a tight transcriptional network is at work to maintain the undifferentiated state of ESCs. Besides, other ESC-specific enhancers that contain binding sites for Oct3/4 and SOX2 have been identified in several genes, including FGF4 (Fibroblast Growth Factor-4), SPP1 (Secreted Phosphoprotein-1), UTF1 (Undifferentiated embryonic cell Transcription Factor-1), FBXO15 (F-box Protein-15), and Lefty1 (Left right determination Factor-1) (Ref.1 & 7). Several other genes containing Oct4/SOX2 binding sites have recently been identified in Mouse. Genes containing 1 Oct4/SOX2 binding site include Lig3 (Ligase-III, DNA, ATP-dependent), KCTD3 (Potassium Channel Tetramerisation Domain containing-3), BIN1 (Bridging Integrator-1), Bmi1 (Bmi1 Polycomb ring finger oncogene) and NASP (Nuclear Autoantigenic Sperm Protein (histone-binding)). Genes containing 5 to 10 sites include InsIG2 (Insulin Induced Gene-2), Ipo11 (Importin-11), MYST4 (MYST Histone Acetyltransferase (monocytic leukemia)-4), NR6A1 (Nuclear Receptor Subfamily-6, group A, Member-1) and STRBP (Spermatid Perinuclear RNA Binding Protein). Genes with greater than 10 sites are POU2F1 (POU domain, class 2, Transcription Factor-1), RANBP17 (RAN Binding Protein-17), and SMYD3 (SET and MYND Domain containing-3). Thirty five genes implicated in chromatin remodeling are correlated to Oct4. Putative positive target genes include SWI/SNF members SMARCC1 (SWI/SNF related, Matrix associated, Actin dependent regulator of chromatin, Subfamily c, member-1), AT rich interactive domain (Swi1 like) containing proteins (ARID domains) ARID1A (AT Rich Interactive Domain-1A (SWI-like)), ARID5B (AT Rich Interactive Domain-5B (SWI-like)), JARID1B (Jumonji, AT Rich Interactive Domain-1B) and JARID2 (Jumonji, AT Rich Interactive Domain-2), which are direct Oct4 target. These ARID domain containing proteins, a subset of the Jumonji C family, have recently been associated with Histone Demethylase activity. Several other genes containing MYST, SET (SET translocation (Myeloid leukemia-associated)), and Chromo, and Bromo domains, which facilitate or recognize specific histone modifications, have also been identified. REST (RE1-Silencing Transcription factor) has been implicated in the repression of neuronal specific genes via its ability to recruit cofactors such as HDACs (Histone Deacetylases), CoREST (Co-repressor of Rest), Sin3, and MECP2 (Methyl CpG binding Protein-2). The identification of REST as a direct Oct4 target, in light of its role in maintaining chromatin plasticity throughout neurogenesis, provides a mechanistic understanding of Oct4’s role in promoting neural differentiation. CoREST and MECP2 are positively and negatively correlated to Oct4 respectively (Ref.8, 9 & 10).
Several members of the TrxG and PcG of transcription factors such as Ash1L (Ash1 (absent, small, or homeotic)-Like (Drosophila)), SUZ12 (Suppressor of Zeste-12 homolog (Drosophila)), ASH2L (Ash2 (absent, small, or homeotic)-Like (Drosophila)), Phc1 (Polyhomeotic-Like 1 (Drosophila)), and Rnf134 (Ring Finger protein-134), Bmi1 (BMI1 Polycomb ring Finger oncogene), and Phc3 (Polyhomeotic-Like 3 (Drosophila))are also correlated to Oct4, with the five latter genes validated as Oct4 targets. Diverse functions for PcG and TrxG genes in cancer, cell cycle control, and stem cell function have recently been described. The direct transcriptional regulation of several members of these complexes places Oct4 central to the coordination of these activities. The localization of SUZ12, a member of PRC2 (Polycomb Repressor Complex-2) at many Oct4 repressed loci in ESC provides indication that Oct4-Polycomb interaction may play a significant role in the active repression of lineage. Oct4 plays an important role in maintaining chromatin structure in Mouse ESC via regulation of and interaction with a unique constellation of PcG and TrxG complexes. While others are positive targets, Bmi1 and Phc3 are found to be negative targets of Oct4. About 38 cell-cycle related genes were positively correlated to Oct4 including CDC25A (Cell Division Cycle 25 homolog-A), GSPT1 (G1 to S Phase Transition-1), PPP1R8 (Protein Phosphatase-1, Regulatory (inhibitor) Subunit-8), CcnB2 (Cyclin-B2), CcnE1 (Cyclin-E1), CcnA2 (Cyclin-A2), CcnB1 (Cyclin-B1), and CcnF (Cyclin-F). Validated Oct4 target CcnF is implicated in cell cycle control at the G1/S and G2/ M checkpoints and has recently been associated with the maintenance of pRb (Retinoblastoma) in a hyperphosphorylated, inactive state. Conversely, the significantly Oct4 correlated PP1 (Protein Phosphatase-1) negative regulatory subunit Nipp1 (PPP1R8) may facilitate the functional inactivation of pRb. Other Cell cycle genes activated by OCT4 include D14Abb1E (DNA segment, Chr 14, Abbott 1 Expressed), IGF2BP1 (Insulin-like Growth Factor 2 mRNA Binding Protein-1) and JARID1B (Jumonji, AT Rich Interactive Domain-1B), which inactivate Rb. Inactivation of pRb is required for self-renewal. An imbalance in either of these processes, possibly emanating from deregulated signaling from the stem cell niche or mutations in the key regulators would lead to unrestrained cellular proliferation(Ref.11 & 12).
Twenty-five apoptotic genes are found to be positively correlated to Oct4, the majority of which,including AATF (Apoptosis Antagonizing Transcription Factor), API5 (Apoptosis Inhibitor-5), Aven (Apoptosis, caspase activation inhibitor), BAG4 (BCL2-Associated athanogene 4), Commd10 (COMM Domain containing-10), Nipa, and Opa1 (Optic Atrophy-1), function to inhibit Apoptosis. In addition, Bin1 (Bridging Integrator-1), Blp1 (BBP-Like Protein-1), Serpinb9 (Serpin peptidase inhibitor, Clade B (ovalbumin), member-9), SH3GLB1 (SH3-domain GRB2-like endophilin-B1), and Casp6 (Caspase6), all apoptosis inducing genes, were found to be negatively correlated to Oct4, with SH3GLB1 and Casp6 confirmed as targets. Thirty genes implicated in DNA damage and repair, are also positively correlated to Oct4. Members of the BASC (Brca1 Associated Surveillance Complex) including BRCA1 (Breast Cancer-1, early onset), Msh2 (MutS Homolog-2, colon cancer, nonpolyposis type 1 (E. coli)), MRE11A (MRE11 meiotic recombination 11 Homolog-A), Rad51 (RAD51 homolog (RecA homolog, E. coli), Blm (Bloom syndrome), Chek1 (CHK1 Checkpoint homolog), as well as PARP1 (Poly (ADP-ribose) polymerase family, member-1), TRp53 (Transformation Related Protein-53), FANCD2 (Fanconi Anemia, Complementation group D2), Tdrd7 (Tudor Domain Containing-7), and Xrcc5 (X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa)) are included. The validation of TRp53, Tdrd7, BRCA1, and PARP1 as direct Oct4 targets strengthens the importance of this group of genes in stem cell function. Tudor domain containing proteins (such as Tdrd7) play an important role in DNA damage response, Casp3 (Caspase3) in skeletal muscle differentiation, and PARP1, TRP53, and BRCA1 helps to modulate differentiation. Tdrd7 is a negative target of OCT4 (Ref.1 & 13).
PML (Promyelocytic Leukemia) and Coil (Cajal Bodies and PML Bodies), Gemin4 (Gem (nuclear organelle) associated Protein-4)and Gemin5 (Gem (nuclear organelle) associated Protein-5) (Gems), Nup35, Nup43, Nup54, Nup98, Nup133, Nup160, Nup188 (Nuclear Pore Complex), Ncl (Nucleolin) and Nolc1 (Nucleolus) and 46 genes implicated in RNA metabolism (Splicing Speckles, Spliceosome, Exosome, and Cajal Bodies) are positively correlated to Oct4. Several genes implicated in nuclear transport such as Ipo11, Kpna1 (Karyopherin Alpha-1 (Importin alpha 5)), Tnpo2 (Transportin-2 (Importin 3, karyopherin beta 2b)) and Tnpo3 (Transportin-3), Xpot (Exportin, tRNA (nuclear export receptor for tRNAs)), Gle1l (GLE1 RNA export mediator-like (yeast)), Xpo5 (Exportin-5) and Xpo6 and direct Oct4 targets IGF2BP1 and Phb were also positively correlated. Oct4 binding functions not only in the transcriptional repression of genes but also presents a means whereby these loci are organized spatially within the nucleus so as to be poised for activation given the appropriate cue. Oct4 promotes self-renewal by inactivating genes which promote differentiation or by activating genes which inhibit differentiation. Most common genes positively regulated by OCT4 include RAR-Alpha (Retinoic Acid Receptor-Alpha), REST, Aqr (Aquarius), JARID2 and SalL4 (Sal-like 4 (Drosophila)). RAR-Alpha, REST and Aqr inhibit Ectoderm and mesoderm differentiation. Negative targets of OCT4 include MEF2A (Myocyte Enhancer Factor-2A) and HoxB1 (Homeo Box-B1). In the initial stages of differentiation HoxB1 is transcriptionally activated which results in chromatin decondensation and reorientation of this locus to the nuclear centre. Finally, about 26 direct Oct4 transcriptional targets have been identified in Mouse ESCs (Ref.14).
In Human ESCs, Oct3/4 has also been reported to directly prevent differentiation towards trophectoderm by interacting with Cdx2 (Caudal type Homeobox-2), a trigger for trophectoderm differentiation, to form a repressor complex. This complex interferes with the autoregulation of these two factors, giving rise to a reciprocal inhibition system that establishes their mutually exclusive expression. As such, the downregulation of Oct3/4 results in an upregulation of Cdx2, and vice versa – a mechanism that might account for the two different pathways that lead to Pluripotent stem cells and to Trophectoderm cells. Oct4 represses gene expression either indirectly by neutralizing activators such as FoxD3 (Forkhead box-D3), or directly by binding to promoters. Oct4 repress the expression of FoxA1 and FoxA2 (Forkhead box-A2) through an interaction with the DNA binding domain of their activator FoxD3. Thus, Oct4 could prevent the differentiation of ESC lineages by acting like a co-repressor of lineage-specific transcription factors like FoxD3. Silencing of Ifn-Tau (Tau Interferon genes) appears to be mediated by Oct4. Oct4 and Ets2 (E26 avian leukemia oncogene 2, 3′ domain) form a complex through interaction between the Oct4 POU domain and the DNA binding domain of Ets2, and as a result quench the transactivation function of Ets2. Oct4 is downregulated, thus, alleviating the co-repression of Ets2 to allow the Trophoectoderm specific genes such as Ifn-Tau to be expressed. Oct4 also binds to a novel PSBP (Pluripotential cell-specific SOX Element-Binding Protein) leading to Nanog gene expression. OCT4 dominantly bound to the Octamer element, while PSBP preferentially bound to the SOX element (Ref.2 & 15).
Recent identification of Oct4 transcriptional targets in ESCs has revealed an unanticipated collaboration between Oct4, SOX2, and Nanog. The identification of common target sites in the regulatory elements of Oct3/4, SOX2 and Nanog has suggested that Oct3/4, SOX2 and Nanog might form a regulatory feedback circuit that maintains pluripotency in human and mouse ES cells; in this circuit, all three transcription factors regulate themselves, as well as each other. Although Oct4 maintain Pluripotency in combination with Nanog and SOX2, even a slight overdose of Oct3/4 triggers differentiation. Oct4 over-expression can inhibit its own expression as well as the expression of SOX2 and Nanog. Recently, a link between Oct4 and tumorigenesis has also been identified. Like ESCs, tumor cells exhibit a unique pattern of gene expression, and thus, may be under the control of one or more master regulators like Oct4. Therefore, an understanding of Oct4 function in Stem cell biology could also lead to novel treatments for certain malignancies (Ref.1 & 16).
References:
1.The human OCT-4 isoforms differ in their ability to confer self-renewal.
Lee J, Kim HK, Rho JY, Han YM, Kim J.
J Biol Chem. 2006 Nov 3;281(44):33554-65.
2.Self-renewal and differentiation of mouse embryonic stem cells as measured by Oct4 expression: the role of the cAMP/PKA pathway.
Faherty S, Fitzgerald A, Keohan M, Quinlan LR
In Vitro Cell Dev Biol Anim. 2007 Jan;43(1):37-47.
3.Oct4 targets regulatory nodes to modulate stem cell function.
Campbell PA, Perez-Iratxeta C, Andrade-Navarro MA, Rudnicki MA.
PLoS ONE. 2007 Jun 20;2(6):e553.
4.Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells.
Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J.
Stem Cells. 2007 Feb;25(2):500-10.
5.Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development.
Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ.
Mol Cell Biol. 2005 May;25(9):3492-505.
6.Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation.
Gu P, LeMenuet D, Chung AC, Mancini M, Wheeler DA, Cooney AJ.
Mol Cell Biol. 2005 Oct;25(19):8507-19.
7.Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation.
Wei F, Scholer HR, Atchison ML.
J Biol Chem. 2007 Jul 20;282(29):21551-60.
8.Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract.
Freberg CT, Dahl JA, Timoskainen S, Collas P.
Mol Biol Cell. 2007 May;18(5):1543-53.
9.Histone deacetylase activity is required for embryonic stem cell differentiation.
Lee JH, Hart SR, Skalnik DG.
Genesis. 2004 Jan;38(1):32-8.
10.Transplanted embryonic stem cells successfully survive, proliferate, and migrate to damaged regions of the mouse brain.
Srivastava AS, Shenouda S, Mishra R, Carrier E.
Stem Cells. 2006 Jul;24(7):1689-94. Epub 2006 Mar 30.
11.Identification of a nuclear localization signal in OCT4 and generation of a dominant negative mutant by its ablation.
Pan G, Qin B, Liu N, Scholer HR, Pei D.
J Biol Chem. 2004 Aug 27;279(35):37013-20.
12.Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers.
Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M.
Genes Dev. 2003 Aug 15;17(16):2048-59.
13.Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells.
Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F.
J Biol Chem. 2005 Feb 18;280(7):5307-17.
14.Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1.
Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B.
Nat Cell Biol. 2006 Oct;8(10):1114-23.
15.Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst.
Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J.
Development. 2005 May;132(9):2093-102.
16.Stimulation of Oct-4 activity by Ewing’s sarcoma protein.
Lee J, Rhee BK, Bae GY, Han YM, Kim J.
Stem Cells. 2005 Jun-Jul;23(6):738-51.
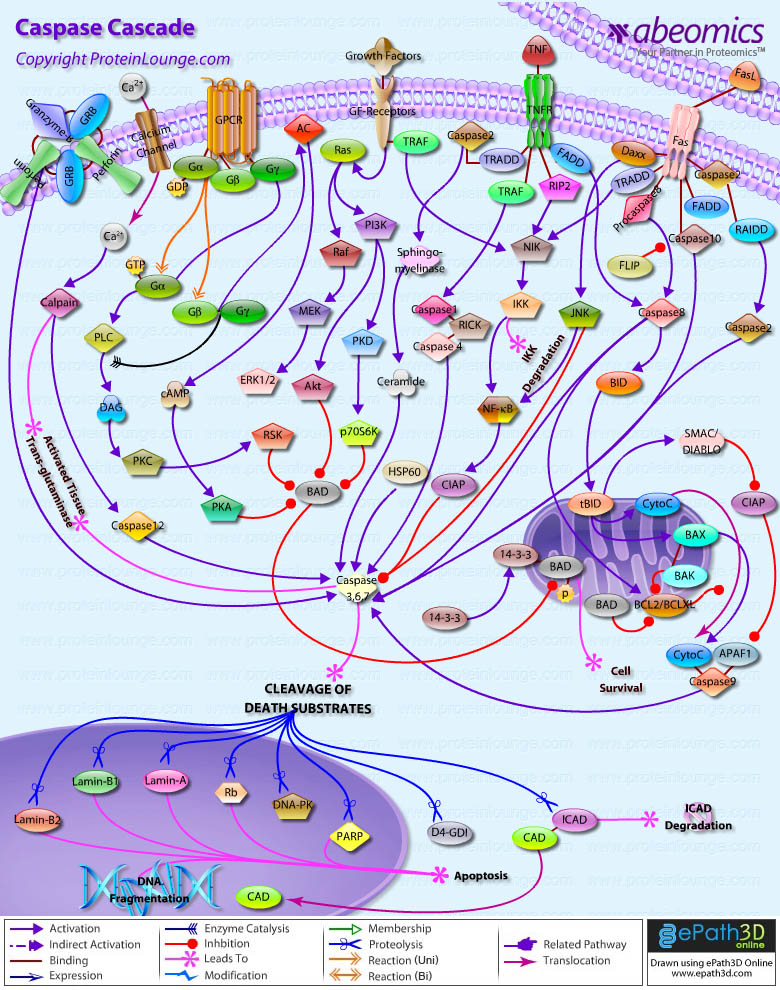
Caspases are a family of cysteine proteases that act in concert in a cascade triggered by apoptosis signaling. The culmination of this cascade is the cleavage of a number of proteins in the cell, followed by cell disassembly, cell death, and, ultimately, the phagocytosis and removal of the cell debris. The Caspase cascade is activated by two distinct routes: one from cell surface and the other from mitochondria (Ref.1). The pathway leading to Caspase activation varies according to the apoptotic stimulus. Initiator Caspases (including 8, 9, 10 and 12) are closely coupled to pro-apototic signals. Pro-apoptotic stimuli include the FasL (Fas Ligand), TNF (Tumor Necrosis Factor), Granzyme-B, GRB (Growth Factor Receptor-Bound Protein), DNA damage, Ca2+ (Calcium) channels and ER (Endoplasmic Reticulum) stress. Once activated, these Caspases cleave and activate downstream effector Caspases (including 3, 6 and 7). Caspase-8 cleaves BID (BH3 Interacting Death Domain). tBID (Truncated BID) disrupts the outer mitochondrial membrane to cause release of the pro-apoptotic factors CytoC (Cytochrome-C) which is crucial for activating pro-Caspase9. CytoC that is released from the intermembrane space binds to APAF1 (Apoptotic Protease Activating Factor-1), which recruits Caspase-9 and in turn can proteolytically activate Caspase-3. SMAC (Second Mitochondria-Derived Activator of Caspase)/DIABLO is also released from the mitochondria along with CytoC during apoptosis, and it functions to promote caspase activation by inhibiting IAP (Inhibitor of Apoptosis) family proteins. ER stress leads to the Ca2+-mediated activation of Caspase-12 (Ref.2).
Fas and the TNFR (TNF Receptor) activate Caspases 8 and 10. Cell death caused by activation of the TNFR or Fas receptors is brought about by the recruitment of the adaptor protein FADD (Fas Associated Death Domain). In the case of the TNFR1, FADD recruitment requires prior binding of TRADD (TNFR-Associated Death Domain Protein). FADD in turn recruits ProCaspase8. The TNFR1 receptor can also mediate activation of Caspase-2 via the recruitment of a death-inducing signaling complex. In this case RIP (Receptor-Interacting Protein) acts as an adaptor for the recruitment of RAIDD (RIP-Associated ICH-1/CED-3-homologous protein with a Death Domain), which subsequently binds to ProCaspase2. TNFR also activates Caspase 3, 6,7 via TRADD, TRAF2 (TNF Receptor-Associated Factor-2) and RICK (RIP-like Interacting Clarp Kinase). TNF not only induces apoptosis by activating Caspase 8 and 10, but can also inhibit apoptosis signaling via NF-KappaB (Nuclear Factor-KappaB), which induces the expression of IAP, an inhibitor of Caspases 3, 7 and 9 (Ref.3).
GRB (Growth Factor Receptor-Bound Protein), Granzyme B and perforin proteins released by cytotoxic T-Cells induce apoptosis in target cells, forming transmembrane pores, and triggering apoptosis, perhaps through cleavage of Caspases, although Caspase-independent mechanisms of Granzyme-B mediated apoptosis have been suggested (Ref.4). After activation, down stream Caspases cleave cytoskeletal and nuclear proteins (structural, signaling proteins or kinases) like PARP (Poly ADP-Ribose Polymerase), DNA-PK (DNA-Dependent Protein Kinase), Rb (Retino Blastoma Tumor Supressor Protein), PAK1 (p21-Activated Kinase-1), GDID4, Fodrin, Lamin-A, Lamin-B1, Lamin-B2, thus inducing apoptosis. Caspase 3 cleaves ICAD (Inhibitor of CAD) to free CAD (Caspase-Activated DNase) to cause DNA fragmentation. The events culminating in Caspase activation and the subsequent disassembly of the cell are the subject of intense study because of their role in many neurodegenerative disorders such as Parkinson’s and Alzheimer’s diseases, autoimmune disorders, and tumorigenesis.
References:
Cell Death Differ. 2002 Sep;9(9):905-14.
Oncogene. 1999 Mar 11;18(10):1781-7.
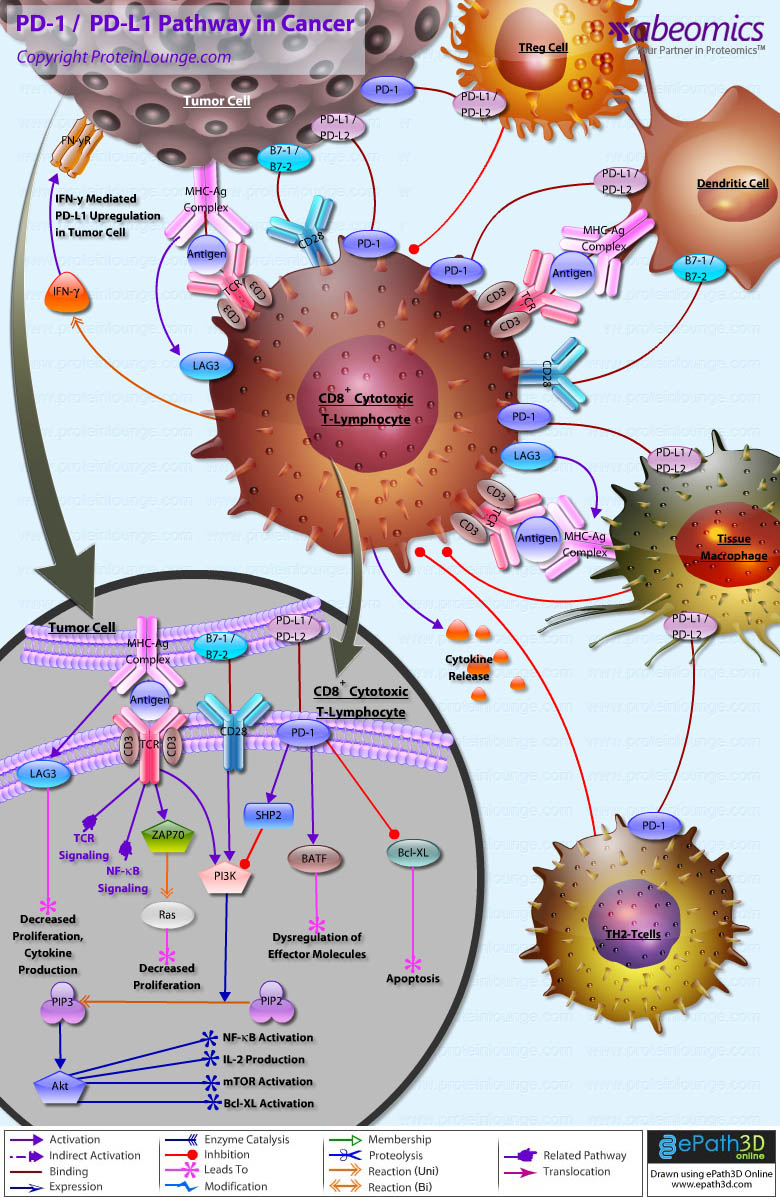
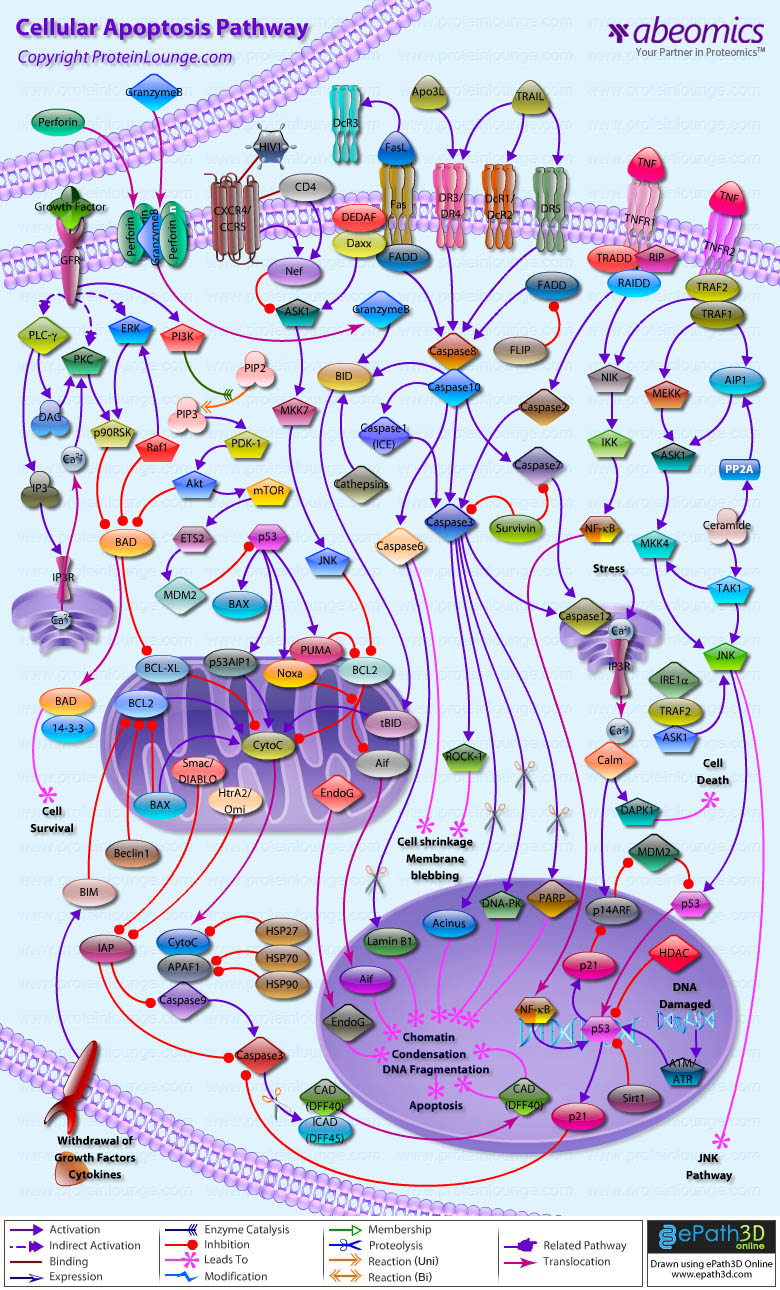
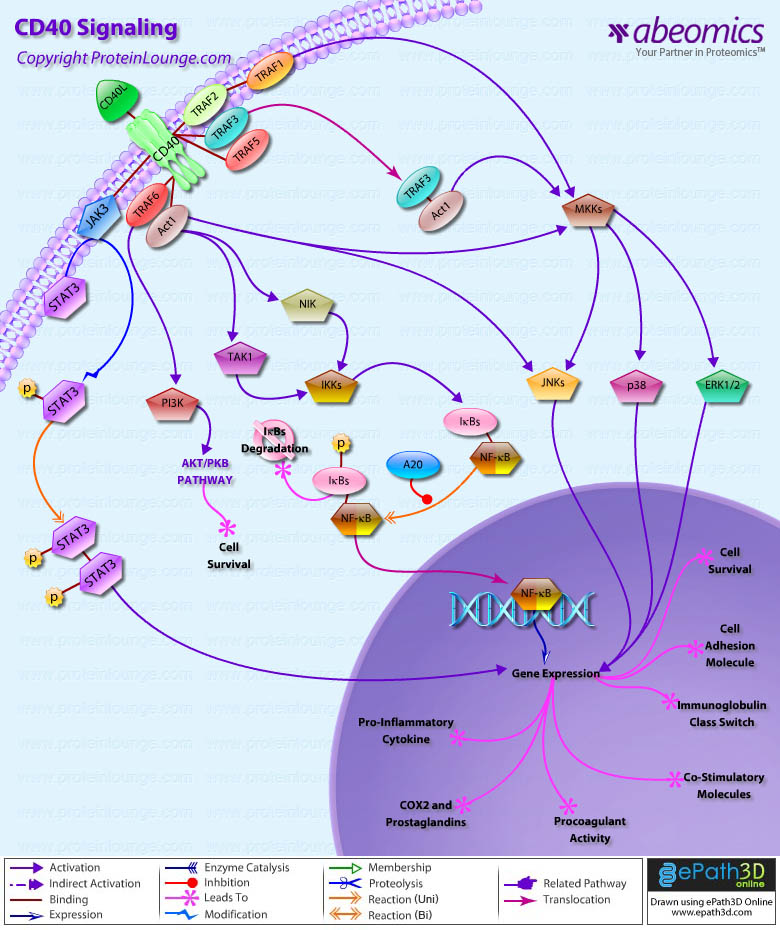
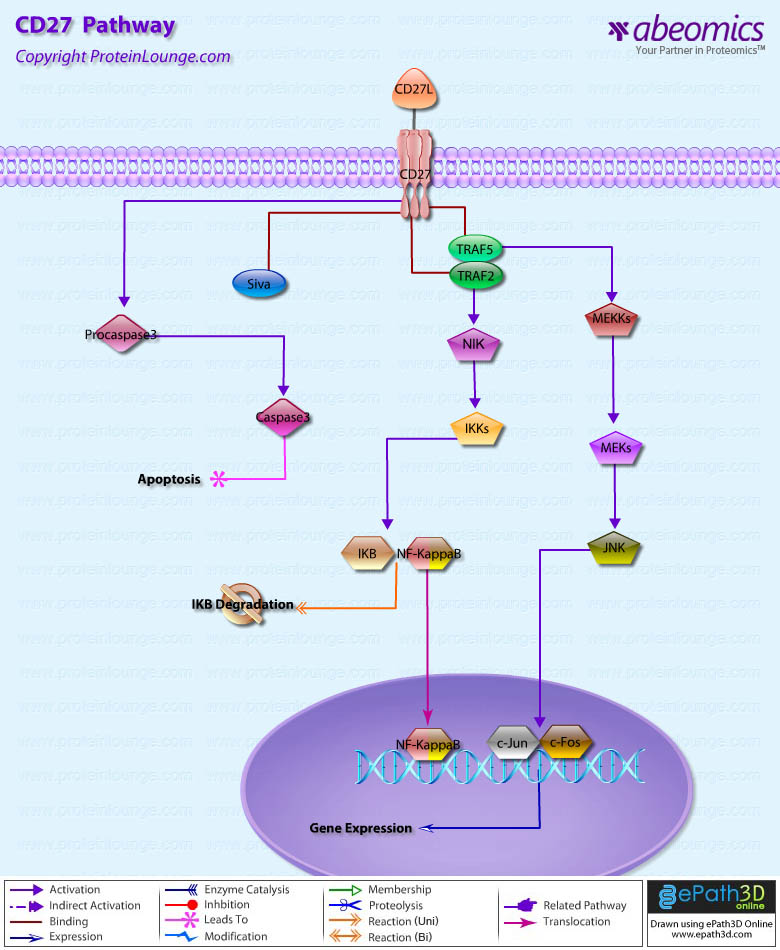
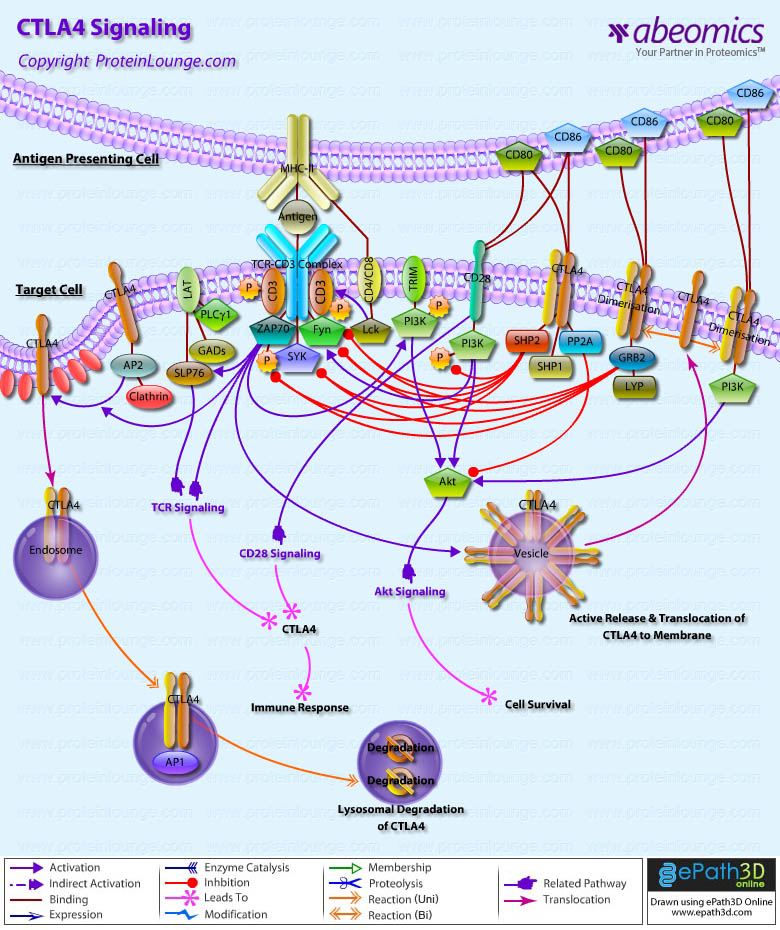
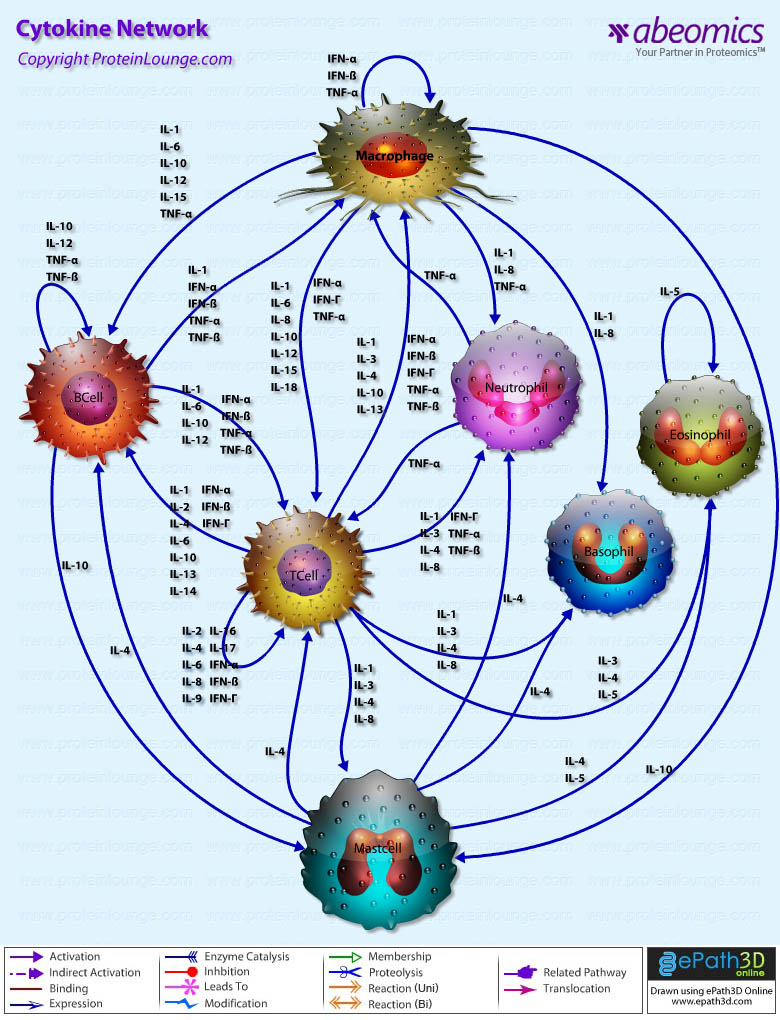
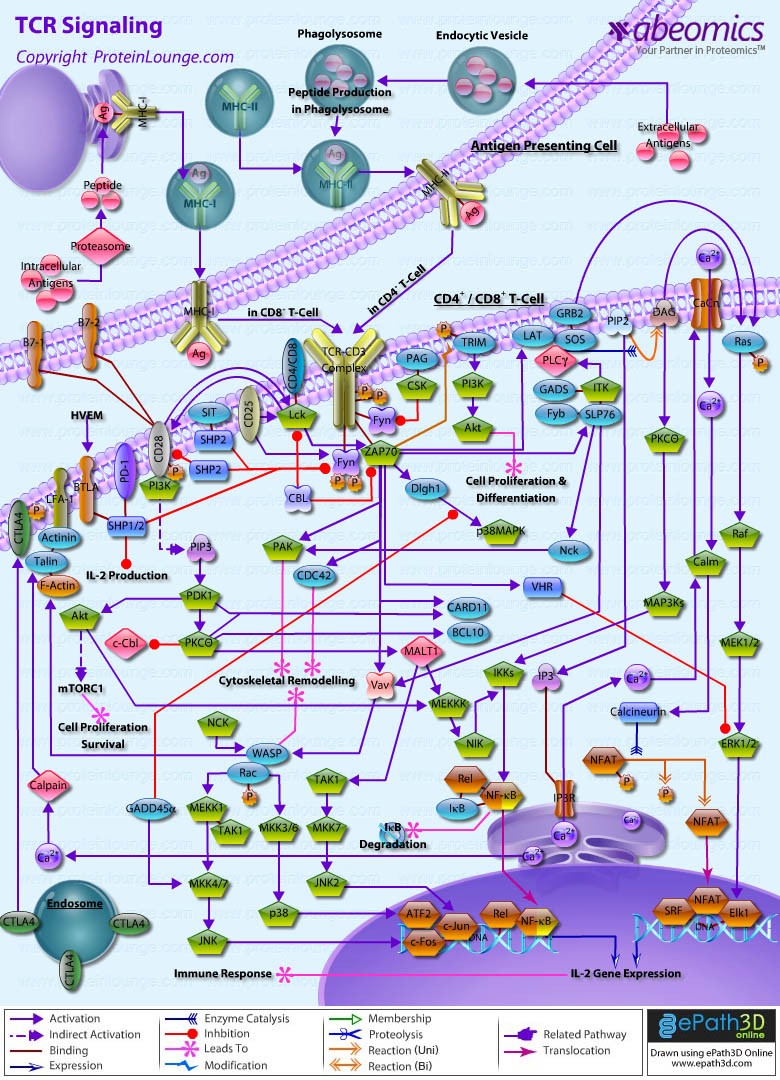
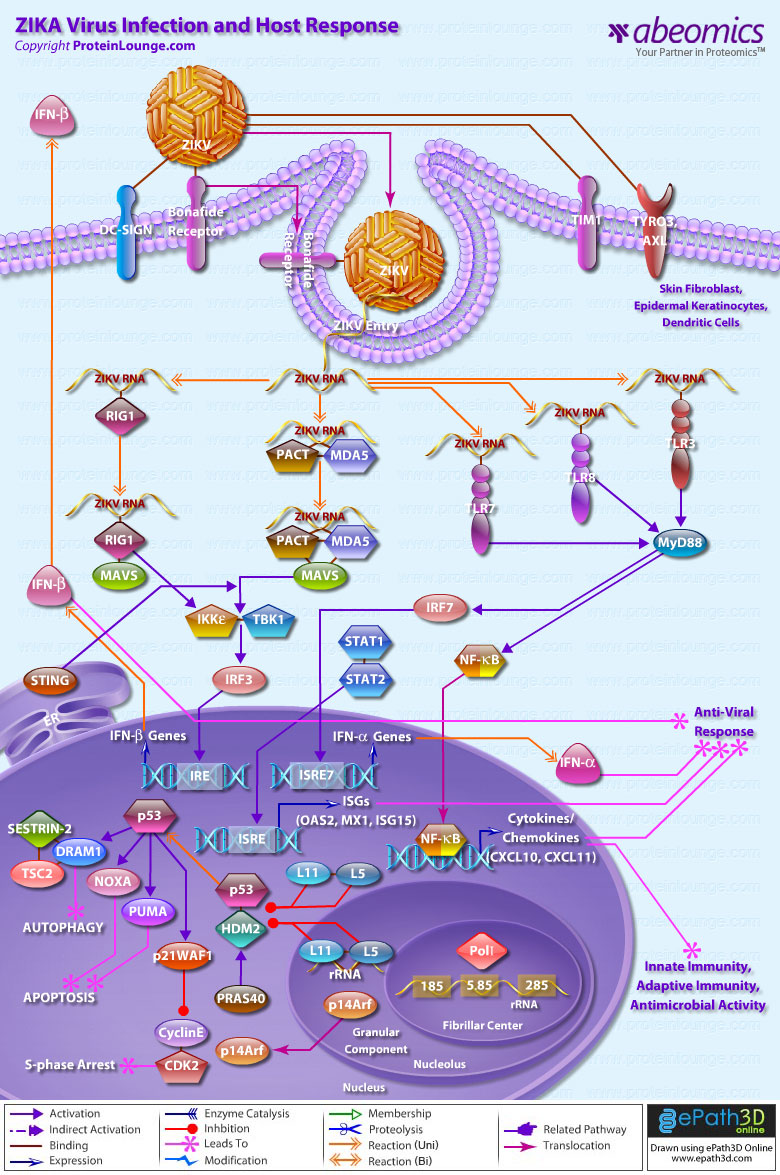

Atopy or Allergic disease is a complex familial disorder with multiple manifestations, including allergic asthma, rhinitis, conjunctivitis, and dermatitis. Allergens are derived from different sources such as cockroaches, ragweed pollens, and house dust mites. The primary immune cell lineages involved in the initiation and progression of allergic inflammation include DCs (Dendritic Cells), mast cells, basophils, eosinophils, and Th2 (Type-2 Helper T) cells. The responses of these principal players in allergic reactions are influenced by the local environments in which they reside. When susceptible or atopic individuals are initially exposed or sensitized to allergens, Antigen-presenting cells capture, process, and present allergen as an allergen-derived peptide fragment in the contest of specific MHC II (HLA II) molecules this induces allergen-specific acquired immune responses, which are characterized by CD4+ T cells that produce a Th2 profile of cytokines for example, IL-4(Interleukin-4), IL-5, IL-9 and IL-13, IL-10 and the presence of allergen-specific IgE. Subsequent challenge with allergen causes the rapid activation of mast cells through allergen and IgE crosslinking, and the release of mediators such as histamine and leukotrienes causes increases in vascular permeability, smooth-muscle contraction and mucus secretion. Late-phase allergic responses are characterized by the additional recruitment and activation of eosinophils and Th2 cells at the site of allergen challenge (Ref.1, 2 & 3).
Mast-cell activation by IgE crosslinking with allergen requires access of allergen into the tissue and input from the adaptive immune system to be effective. Mast-cell activation requires not only the synthesis of specific IgE by B cells (regulated by interleukin-4 (IL-4) and IL-13 derived from Th2 cells and basophils), but also mast-cell priming by IL-4 for enhanced mediator release. The subsequent release of mast-cell mediators such as histamine, LTC4 (Leukotriene C4) and PGD2 (Prostaglandin D2) leads to an early reaction, consisting classically of a ‘wheal and flare’ reaction of the skin or the mucosa. These mediators affect the mucosa, the blood vessels and sensory nerves (pain). Other mast-cell mediators, such as IL-3, IL-5, IL-8,IL-33, TNF (Tumor-Necrosis Factor),NT3 (Neurotrophin 3) and proteases contribute to the initiation of a facultative latephase reaction by recruiting and activating eosinophils, neutrophils and Th2 cells, and by interaction with tissue cells such as nerve cells, smooth-muscle cells, endothelial cells and the epithelium. Ongoing dysregulation of such cell types not only causes symptoms of allergy, but also organ dysfunction, including loss of barrier function and, subsequently, increased bacterial translocation. This enables non-specific triggers to access mast cells, dendritic cells and other cells.Triggers such as bacterial products, or immunoglobulin such as monomeric IgE and light chains might perpetuate the inflammatory process, even in the absence of allergen (Ref.4 & 5). Eotaxin is mainly produced by epithelial cells, but in some conditions it is also produced by other cell types, such as mast cells and alveolar macrophages. Signals that trigger the production of eotaxin by epithelial cells come from lymphocytes, but possibly also mast cells and dendritic cells. Eotaxin recruits Th2 cells that in turn produce IL-4 and IL-5 and amplify all the effects on epithelial cells and mast cells, resulting in the production of more eotaxin. Significant levels of eotaxin result in eosinophil recruitment and degranulation, further Th2 recruitment, basophil degranulation and mast cell migration and differentiation. Antigen-activated basophils secrete soluble factors that stimulate tissue-resident nonhaematopoietic cells, such as fibroblasts, to produce various chemokines that in turn recruit inflammatory cells, including eosinophils and neutrophils, to the skin lesions. These results suggested that basophils have an important and nonredundant role in chronic allergic inflammation as initiators rather than effectors of the inflammatory response (Ref.6, 7 & 8).
Once attracted to the site of inflammation the eosinophil becomes activated and, as a result of this, secretes several tissue-toxic mediators. These are either basic or granule-stored proteins, ECP (Eosinophil Cationic Protein), EPO (Eosinophil Peroxidase), eosinophil protein/eosinophil-derived neurotoxin, and MBP (Major Basic Protein) or reactive oxygenfree radicals. The eosinophil also produces a wide array of different cytokines, leukotrienes, chemokines, and lipid mediators, granulocyte/macrophage colony-stimulating factor, and is therefore, in addition to being an effector cell, thought to play an immunoregulatory role in inflammatory processes as well as taking part in tissue remodeling. EDN (Eosinophil-derived neurotoxin), a member of the RNase A superfamily, is a mediator produced by human eosinophils and placental epithelial cells. In addition to its ribonuclease activity EDN reduces the infectivity of respiratory syncytial virus for target cells in vitro and is also responsible in part for the anti HIV-1 activity found in the supernatants of mixed lymphocyte cultures by identifying EDN (Ref.9, 10 & 11). TSLP emerged as a central player in the development of allergic symptoms, especially in the airways, and is a prime regulatory cytokine at the interface of virus- or antigen-exposed epithelial cells and DCs. DCs activated by epithelium-derived TSLP can promote naïve CD4+ T cells to adopt a Th2 phenotype, which in turn recruit eosinophilic and basophilic granulocytes as well as mast cells into the airway mucosa. These different cells secrete inflammatory cytokines and chemokines operative in inducing an allergic inflammation and atopic asthma. TSLP is, thus, involved in the control of both an innate and an adaptive immune response (Ref.12).
1.Regulation of allergic inflammation and eosinophil recruitment in mice lacking the transcription factor NFAT1: role of interleukin-4 (IL-4) and IL-5.
Viola JP, Kiani A, Bozza PT, Rao A.
Blood. 1998 Apr 1;91(7):2223-30.
2.The influence of TSLP on the allergic response.
Comeau MR, Ziegler SF.
Mucosal Immunol. 2010 Mar;3(2):138-47. doi: 10.1038/mi.2009.134.
3.Identification of antigenic epitopes on human allergens: studies with HLA transgenic mice.
Chapoval SP, David CS.
Environ Health Perspect. 2003 Feb;111(2):245-50.
4.Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data.
Bischoff SC.
Nat Rev Immunol. 2007 Feb;7(2):93-104.
5.Potential role of interleukin-10-secreting regulatory T cells in allergy and asthma.
Hawrylowicz CM, O’Garra A.
Nat Rev Immunol. 2005 Apr;5(4):271-83.
6.Eotaxin: from an eosinophilic chemokine to a major regulator of allergic reactions.
Gutierrez-Ramos JC, Lloyd C, Gonzalo JA.
Immunol Today. 1999 Nov;20(11):500-4.
7.Improvement of cellulase activity using error-prone rolling circle amplification and site-directed mutagenesis.
Vu VH, Kim K.
J Microbiol Biotechnol. 2012 May;22(5):607-13.
8.Th2 cell-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation.
Cameron L, Webster RB, Strempel JM, Kiesler P, Kabesch M, Ramachandran H, Yu L, Stern DA, Graves PE, Lohman IC, Wright AL, Halonen M, Klimecki WT, Vercelli D.
J Immunol. 2006 Dec 15;177(12):8633-42.
9.Eosinophil cationic protein (ECP) is processed during secretion.
Woschnagg C, Rubin J, Venge P.
J Immunol. 2009 Sep 15;183(6):3949-54. doi: 10.4049/jimmunol.0900509.
10.Eosinophil-derived neurotoxin acts as an alarmin to activate the TLR2-MyD88 signal pathway in dendritic cells and enhances Th2 immune responses.
Yang D, Chen Q, Su SB, Zhang P, Kurosaka K, Caspi RR, Michalek SM, Rosenberg HF, Zhang N, Oppenheim JJ.
J Exp Med. 2008 Jan 21;205(1):79-90. doi: 10.1084/jem.20062027.
11.Inhibitory effects of ketotifen on eotaxin-dependent activation of eosinophils: consequences for allergic eye diseases.
Woerly G, Loiseau S, Loyens M, Schoch C, Capron M.
Allergy. 2003 May;58(5):397-406.
12.Signal transduction around thymic stromal lymphopoietin (TSLP) in atopic asthma.
Sebastian K, Borowski A, Kuepper M, Friedrich K.
Cell Commun Signal. 2008 Aug 25;6:5. doi: 10.1186/1478-811X-6-5.
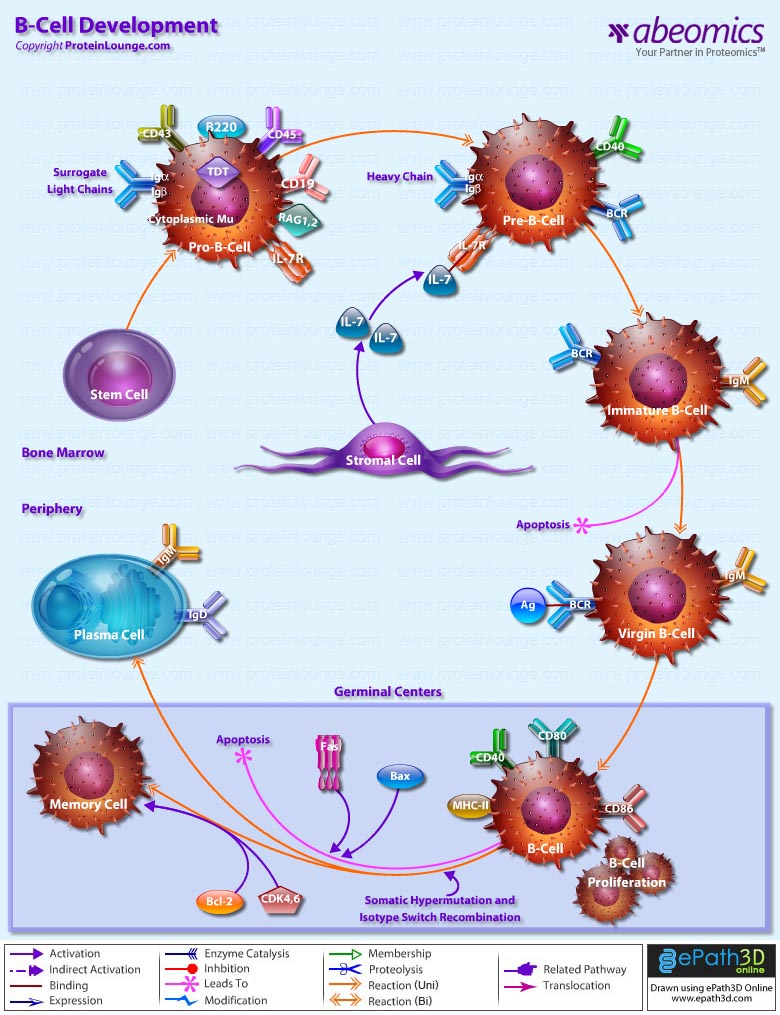
B lymphocytes are a population of white blood cells that express clonally diverse cell surface Ig (immunoglobulin) receptors, recognizing specific antigenic epitopes. The development of B-cells encompasses a continuum of stages that begin in primary lymphoid tissue (eg., human fetal liver and fetal/adult marrow), with subsequent functional maturation in secondary lymphoid tissue (eg., human lymph nodes and spleen). The functional/protective end point is antibody production by terminally differentiated plasma cells (Ref.1 ). B cells express immunoglobulin (Ig) molecules on their outer surface and secrete them into the extracellular space. Secreted Ig is known as antibody. Antibodies serve as effector molecules that neutralize microbes by binding to exposed antigens and targeting them to other components of the immune system, such as phagocytic cells and complement, that effect clearance. The genes that encode for antibodies are generated by many diversifying mechanisms including combinatorial rearrangement of gene segments, addition of non-templated (n) nucleotides at the junctions, and somatic hyper-mutation. Ig genes generate diversity in two stages: an antigen-independent and an antigen-dependent stage. Antigen independent diversity is generated in the bone marrow, where B cells originate, by combinatorial rearrangement of gene segments and junctional diversity. Combinatorial diversity is created in a number of ways. First, each antibody molecule comprises one heavy- and one light-chain protein. Both the light- and heavy-chain genes are encoded by gene segments that are genetically rearranged during a process known as V(D)J recombination. Heavy chains are made up of three gene segments—variable (VH), diversity (DH) and joining (JH) where as light chains only have a V and J segment. In humans, there are ~50 known functional VH segments, 27 known functional DH segments, and six known functional JH segments. This arrangement allows for~8100 combinations in the heavy chain alone. Humans also have two light-chain loci, κ and λ. Only one of these loci is expressed per cell so that each antibody either has a κ light chain or a λ light chain. Humans have 44 functional V κ, 5 Jκ, 33Vλ genes and 5 Jλ resulting in 220 possible κ chains and 165 possible λ chains. Thus this combinatorial rearrangement alone allows for greater than 3 million antibodies. Junctional diversity is the result of multiple recombination site choices for each recombination event and the addition of n nucleotides. n nucleotides are sometimes added at the junction by terminal deoxynucleotidyl transferase (TdT) between adjoining gene segments. Antigen-dependent diversity is generated by somatic hyper mutation in the periphery in a manner dependent on activation-induced cytidine deaminase (AID); during this process, mutations in the Ig genes are accumulated at rate of up to 106 times the normal background rate B cells are subsequently selected for enhanced affinity for the eliciting antigen. It is estimated that these processes of diversification can generate ~1012 different antibodies making it challenging to correctly identify the underlying germline gene segments and subsequently the sequences of the complementarity determining regions (CDRs).(Ref. 2 ).
B cell development starts in bone marrow with the commitment of hematopoietic stem cells (HSCs) to the B cell lineage and ends with formation of mature B cells in peripheral secondary lymphoid organs (e.g., the spleen).It is the sequential expression and assembly of the components of the B cell antigen receptor (BCR) which defines each developmental stage. The first developmental stage exhibiting commitment to the B cell lineage is called pro-B and is characterized by rearrangement of the Ig heavy chain. In the pro-B stage Igα (CD79a) and Igβ (CD79b) are expressed at the cell surface in association with chaperon proteins such as calnexin. As soon as the heavy chain (µH) is successfully recombined, it is assembled with Igα and Igβ and the surrogate light chains (SCLs) λ5 and VpreB to form the pre-BCR like IgM (Ref.3 & 4 , ).Surface expression of this receptor marks the transition to the pre-B stage. Marrow stromal cell–derived interleukin-7 (IL-7) is a non-redundant cytokine for B-cell development that promotes V to DJ rearrangement and transmits survival/proliferation signals (Ref.1 18725575). Unsuccessful recombination (with heavy chain VDJ fragments that are not in a proper reading frame) or unsuccessful pairing of the pre-BCR components results in pro-B cells that are unable to proceed to the pre-B stage and leads to apoptosis. In the pre-B stage, the light chain V and J fragments are recombined, and, again, successful assembly of the mature form of the BCR marks the transition to the immature stage. In addition to their VDJ recombination status, all pro-B and pre-B stages can be recognized by their patterns of surface markers expression and their proliferative state. Because DNA replication during proliferation is associated with homologous recombination and VDJ rearrangement is associated with non homologous recombination, these two processes are mutually exclusive. Pro-B and pre-B stages are thus characterized by waves of VDJ recombination followed by waves of proliferation. To achieve this pattern, the expression of the two main enzymes responsible for VDJ rearrangement, the recombinase associated genes 1 and 2 (RAG1 and RAG2), are tightly regulated during the cell cycle; they are highly active in G0 and degraded before entrance into S phase. The developing B cell thereby ensures that no events of non-homologous recombination will occur during DNA replication and avoids an increase in the mutation rate (Ref.3 ).
At this point of development and before leaving for periphery, alternative mRNA splicing of the heavy chain gene transcript produces IgD, as well as IgM, as membrane bound Ig. Such B-Cells are called naïve or Virgin B-Cells because they have yet to be exposed to their specific antigen (Ref. 5 ). Transition to the mature stage happens if the BCR of the immature/transitional B cell does not find its cognate antigen after several days of bone marrow and peripheral trafficking. But once encountering an antigen, B cells become activated and differentiate into memory B cells or plasma cells. These dynamic processes occur in the germinal center (GC) and extrafollicular areas (Ref. 3 ).After encountering antigen, B cells become activated and differentiate into memory B cells or plasma cells. These dynamic processes occur in the germinal center (GC) and extrafollicular areas. GC is critically important to B-cell immune responses, in particular in T-dependent responses to protein antigens. In the GC, B cells proliferate rapidly and can undergo isotype switching and somatic hypermutation (SHM). SHM is thought to be critical for selecting B cells with increasing BCR (or antibody) affinity and specificity for the immunizing antigen, and these B cells can be differentiated into memory B cells or plasma cells (Ref.6 ).
LeBien TW, Tedder TF.
Blood. 2008 Sep 1;112(5):1570-80.
2.SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements.
Munshaw S, Kepler TB.
Bioinformatics. 2010 Apr 1;26(7):867-72.
3.Signaling proteins and transcription factors in normal and malignant early B cell development.
Pérez-Vera P, Reyes-León A, Fuentes-Pananá EM.
Bone Marrow Res. 2011;2011:502751.
4.Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells.
Melchers F, ten Boekel E, Seidl T, Kong XC, Yamagami T, Onishi K, Shimizu T, Rolink AG, Andersson J.
Immunol Rev. 2000 Jun;175:33-46.
5.A B-cell receptor-specific selection step governs immature to mature B cell differentiation.
Levine MH, Haberman AM, Sant’Angelo DB, Hannum LG, Cancro MP, Janeway CA Jr, Shlomchik MJ.
Proc Natl Acad Sci U S A. 2000 Mar 14;97(6):2743-8.
6.B-lymphocyte biology.Kurosaki T.
Immunol Rev. 2010 Sep;237(1):5-9.
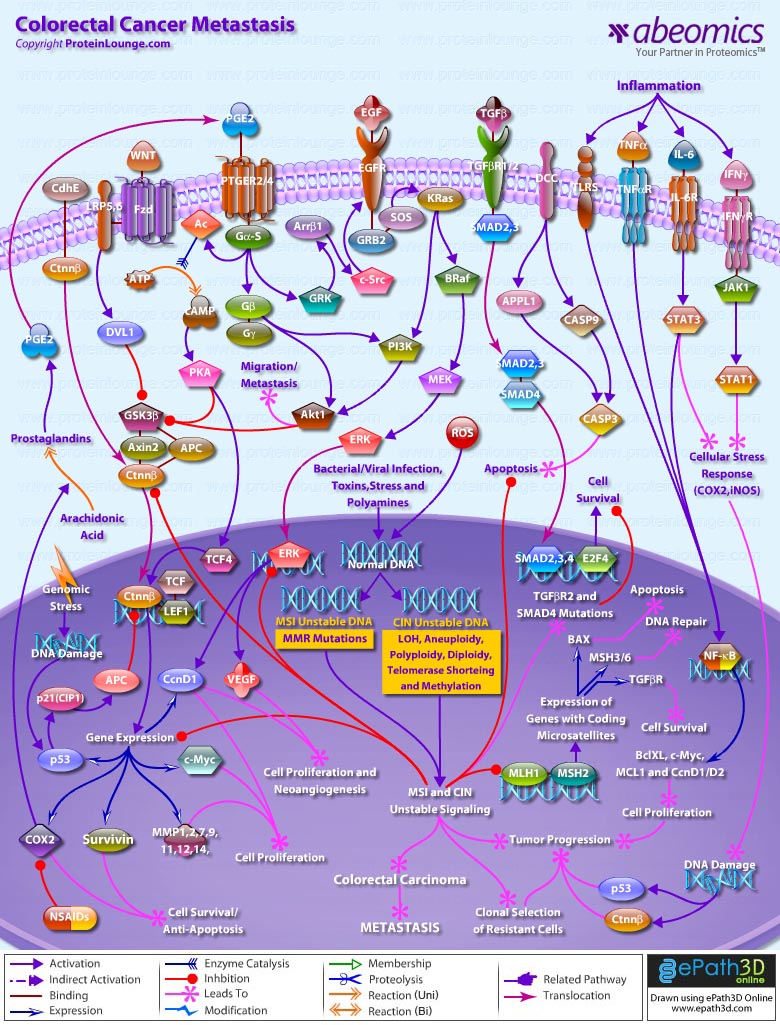
Colorectal cancer represents a relatively well-characterized tumorigenesis paradigm and colorectal carcinoma is one of the leading causes of cancer-related death. Colorectal cancer results from the accumulation of genetic alterations. Genomic instability creates a permissive state in which a potential cancer cell is allowed to acquire enough mutations to become a cancer cell. Several forms of genomic instability are common in colon cancer: MSI (Microsatellite Instability), CIN (Chromosome Instability), and chromosome translocations. MSI occurs in approximately 15% of colon cancers and results from inactivation of the MMR (Mismatch Repair) system secondary to either MMR gene mutations or hypermethylation of MMR gene promoters. It promotes tumorigenesis by generating mutations in target genes that possess coding microsatellite repeats. CIN occurs in most other colon cancers and leads to a different pattern of gene alterations that culminate in tumor formation. It seems to result from mutations in genes that control mitosis, DNA damage repair, centrosome structure and function, and other fundamental processes in DNA replication (Ref.1 & 2). In general the genetic alterations in colorectal cancer progression are determined by one of two separate and distinct underlying pathways of genomic instability. The first pathway, CNI, is characterized by allelic losses, LOH (Loss of Heterozygosity), aneuploidy, diploidy, polyploidy, telomere shortening and methylation. The second pathway, MSI, is characterized by an abundance of subtle DNA mutations. Although the genes causing chromosomal instability remain unknown, MSI is caused by inactivation of a DNA mismatch repair gene (predominantly MLH1 (MutL Homolog-1) or MSH2. Major environmental factors, which influence such gene alternations, include invasion by pathogens (bacterial and viral infections), toxins, stress, polyamines, inflammation, and generation of ROS (Reactive Oxygen Species). The MSI and CIN mutations mainly alter the WNT (Wingless-Type MMTV Integration Site Family), CdhE (Epithelial Cadherin), Prostaglandin, EGFR (Epidermal Growth Factor Receptor), TGF-BetaR (Transforming Growth Factor-Beta Receptor) and DCC (Deleted in Colorectal Carcinoma) signaling routes to facilitate proliferation of colorectal carcinoma. These signaling routes act as check gates in the development of human neoplasia (Ref.3 & 4).
Conceptually, a gatekeeper in human neoplasia refers to a gene that controls the initiation (or is rate-limiting) of a neoplasm in a specific tissue. That is to say, the development of an early neoplasm in this tissue is almost universally associated with (and possibly requires) alteration of this target gene. In contrast, a caretaker refers to a gene that controls the rate of accumulation of genetic alterations (or mutations) during neoplastic progression. Mutation and subsequent inactivation of the APC (Adenomatous Polyposis Coli) tumor suppressor gene in colorectal neoplasia best exemplifies the gatekeeper model. Although APC has a number of putative functions, but the most ascribe role in intestinal neoplasia is to control the WNT signaling pathway via regulation of Ctnn-Beta (Catenin-Beta) levels. In normal cells, the WNT ligands signal via seven transmembrane Fzd (Frizzled) family receptors together with LRP5 (Lipoprotein Receptor-related Protein-5) and LRP6 co-receptors transduce signals through Dsh (Dishevelled) to inhibit GSK3Beta (Glycogen Synthase Kinase-3-Beta) action. In the absence of WNT signaling, Ctnn-Beta is associated with a cytoplasmic complex containing GSK3Beta (Glycogen Synthase Kinase-3-Beta), AXIN (Axis Inhibitor) and the APC (Adenomatous Polyposis Coli) protein, which leads to the degradation of Ctnn-Beta by Proteosome. However, in the presence of WNT signaling, Dsh enhances the phosphorylation of GSK3Beta, which inhibits the ability of GSK3Beta leading to accumulation of free and unphosphorylated Ctnn-Beta in the cytoplasm, which then translocates to the nucleus and associates with TCF and LEF to promote cell survival (anti-apoptosis) and cell proliferation. CdhE also triggers Ctnn-Beta translocation to the nucleus. APC inactivation in tumors by MSI and CIN mutations leads to Ctnn-Beta stabilization, activation of the TCF (Transcription Factor)/LEF (Lymphoid Enhancer-Binding Factor) transcription factor, and subsequent up-regulation of c-Myc, CcnD1 (Cyclin-D1), COX2 (Cyclooxygenase-2), Survivin, p53 and MMP (Matrix Metalloproteinase)-1/2/7/9/11/12/14 (Ref.3,4 & 5). Also genomic stress conditions that induce DNA damage and subsequent p53 activation, leads to the inactivation of p53—>p21(CIP1) (Cyclin Dependent Kinase Inhibitor-p21)—>APC signaling route to up-regulate Ctnn-Beta induced gene transcription and tumor proliferation. Ctnn-Beta, like APC acts as a key mediator in WNT regulation of multiple cellular functions in embryogenesis and tumorigenesis (Ref.6).
Similarly, disruption of COX2 gene (which encodes the key enzyme for conversion of Arachidonic acid to Prostaglandins) and its derived PGE2 (Prostaglandin-E2); this conversion is checked by NSAIDs (Non-Steroidal Anti-Inflammatory Drugs) that inhibit COX2 action), stimulate the growth of cancer cells and promote tumor angiogenesis. PGE2 signals via PTGER2 (Prostaglandin-E Receptor-2) and PTGER4 and activates the Ctnn-Beta/TCF-dependent transcription in colon cancer cells through GN-AlphaS (GN-AlphaS Complex Locus)—>AC (Adenylate Cyclase)—>cAMP (Cyclic Adenosine 3′,5′-monophosphate)—>PKA (Protein Kinase-A) pathway. Mechanistically, PGE2 increase the phosphorylation of GSK3Beta and consequently accumulate Ctnn-Beta. In addition, PGE2 induces the expression of TCF4 transcription factor, which forms transcriptionally active complex with Ctnn-Beta (Ref.7). In a parallel cascade the free G-protein Beta and Gamma subunits activate PI3K (Phosphatidylinositde-3 Kinase) and Akt1 (v-Akt Murine Thymoma Viral Oncogene Homolog-1) leading to the phosphorylation of GSK3Beta and the anticipated activation of Ctnn-Beta. Importantly, this pathway seems to be only partially responsible for PGE2-induced Ctnn-Beta-dependent transcriptional activity (Ref.1). Moreover, COX2-derived PGE2 also provides growth advantage to colorectal carcinomas through transactivation of the EGF (Epidermal Growth Factor)/EGFR (Epidermal Growth Factor Receptor) signaling system. When COX2-derived PGE2 binds to and activates the PTGER4, it induces the phosphorylation of the receptor by GRKs (G-Protein-Dependent Receptor Kinases). The phosphorylation on intracellular loops of the PTGER4 attracts Ar-Beta1 (Beta-Arrestin-1). The translocation of Ar-Beta1 to the receptor triggers the dephosphorylation of Ar-Beta1 at Ser14 (Serine-412), thus allowing its association with c-Src. Ar-Beta1 activates c-Src, which then initiates the transactivation of the EGFR and subsequent downstream signaling through Akt1. The EGFR/SOS (Son of Sevenless)/GRB2 (Growth Factor Receptor-Bound Protein-2)—>PI3K—>Akt1 and EGFR/SOS/GRB2—>KRas (Kirsten-Ras)—>PI3K—>Akt1 signaling cascades are involved in the migration and metastasis of colorectal carcinomas (Ref.8). Whereas, activation of the EGFR/SOS/GRB2—>KRas—>BRaf (v-Raf Murine Sarcoma Viral Oncogene Homolog-B1)—>MEK (MAPK/ERK Kinase)—>ERK (Extracellular Signal-Regulated Kinase) signaling cascade is vital for PGE2 induced gene expression of CcnD1 and VEGF (Vascular Endothelial Growth Factor) that plays critical roles in cell proliferation and neoangiogenesis and is vital for establishment of colorectal carcinogenesis as PGE2 and mutated Ctnn-Beta (via MSI/CIN) stimulate the transcription of CcnD1 and VEGF in a synergistic fashion. This indicates that COX2/PGE2 induced EGFR signaling promote the growth of colon cancers through enhancement of tumor angiogenesis (Ref.7).
Apart from the gatekeeper genes, the caretaker genes are also equally important during the manifestation of colon cancers. In this regard, the DNA mismatch repair genes serve as the prototypic model of caretaker genes in human neoplasia (Ref.3). Among the common targets are MLH1 and MSH2 as they control the transcription of genes having coding microsatellite sequences like TGF-BetaR (that promotes cell survival); MSH3/6 (responsible for DNA repair) and BAX (Bcl2 Associated-X Protein) (enhance apoptosis). Signaling by TGF-Beta (Transforming Growth Factor-Beta) superfamily members is disrupted in various intestinal hyperproliferative states (TGF-Beta/TGF-BetaR—>SMAD (Sma and MAD (Mothers Against Decapentaplegic) Related Protein)2/3/4—>E2F4 (E2F Transcription Factor-4)—>cell survival). In the colorectal carcinogenetic sequence, inactivation and mutations of TGF-Beta signaling components occur at the adenoma to carcinoma transition. The most common mutations inactivate TGF-BetaR2, which accounts for the majority of MSI tumors and approximately 55% of MSS (Microsatellite Stable) tumors (Ref.9 & 10). The MSI and CIN mutations are important for clonal selection of resistant cells and tumor progression. Further loss of chromosome 18q21 is well documented in colorectal cancer, and it is suggested that this loss targets the DCC (Deleted in Colorectal Carcinoma), SMAD4 and SMAD2 genes. DCC drives cell death independently of both the mitochondria-dependent pathway and the Death Receptor/Caspase-8 pathway. Moreover, DCC interacts with APPL (Adaptor Protein containing pH domain, PTB domain and Leucine Zipper Motif-1)/DIP13Alpha, Caspase3 and Caspase9 and drives the activation of Caspase3 through Caspase9 without a requirement for CytoC (Cytochrome-C) or APAF1 (Apoptotic Protease Activating Factor-1). DCC acts as a putative tumor suppressor. The importance of SMAD4, a downstream regulator in the TGF-Beta signaling pathway, in colorectal cancer is associated with late-stage or metastatic disease state. SMAD4 mutations occur before colorectal cancers become aneuploid/polyploid, but after the MSI+ and MSI- pathways diverge. MSI+ cancers may diverge first, followed by CIN+ cancers, leaving other cancers to follow a CIN-MSI- pathway (Ref.11, 12 & 13).
Along with MSI and CIN pathways, the inflammatory bowel diseases (IBDs), such as UC (Ulcerative Colitis) and Crohn’s disease, are well known to increase the risk of developing colorectal cancer. Tumor progression is caused by pro-inflammatory cytokines like Ifn-Gamma (Interferon-Gamma), IL-6 (Interleukin-6), TNF-Alpha (Tumor Necrosis Factor-Alpha). Inflammation induced activation of TNF-Alpha/TNF-AlphaR (Tumor Necrosis Factor-Alpha Receptor) and TLRs (Toll-Like Receptors) activate NF-KappaB (Nuclear Factor-KappaB) induced gene activation; whereas, IL-6/IL-6R (Interleukin-6 Receptor) and IFN-Gamma/IFN-GammaR (Interferon-Gamma-Receptor) activate STAT3 (Signal Transducer and Activator of Transcription-3) and STAT1 (through JAK2 (Janus Kinase-2)), respectively. Aberrantly activated STAT3 and NF-KappaB signals participate in oncogenesis through up-regulation of genes such as apoptosis inhibitors (BclXL (Bcl2 Related Protein Long Isoform), MCL1 (Myeloid Cell Leukemia-1)) and cell cycle regulators (c-Myc, CcnD1/D2) to induce cell proliferation and anti-apoptotic factors that leads to tumor progression. Aberrantly activated STAT1 and STAT3 signal induces cellular stress responses (up-regulation COX2, iNOS (Inducible Nitric Oxide Synthase)) and leads to DNA damage (activating p53, Ctnn-Beta), resulting in clonal selection of resistant cells. These signals then orchestrate tumor formation in the colon (Ref.14 & 15). With the many changes that are taking place in dietary habits and lifestyle, colorectal cancer has become a global concern, for which novel treatment needs to be developed in order to add to the therapeutic armamentarium. The development of the RNAi (RNA interference) technology promises new modality of treatment, which may be used in combination with the existing therapies. RNAi is a sequence-specific post-transcriptional gene silencing mechanism, which is triggered by dsRNA (double-stranded RNA) and causes degradation of mRNA homologous in sequence to the dsRNA. This new approach has been successfully adopted to inhibit virus replication and tumorigenicity. DNA vector-based strategies for delivery of siRNA (small interfering RNA) into mammalian cells, further expands the utility of RNAi. siRNAs may be the best tools for target validation in biomedical research and cancer therapeutics, because of their exquisite specificity, efficiency and endurance of gene-specific silencing. Therefore, RNAi provides the opportunity to pursue an exciting new therapeutic approach to treat colorectal cancer (Ref.16).
Liu X, Lazenby AJ, Siegal GP.
Adv. Anat. Pathol. 2006 Sep;13(5):270-274.2.Genomic instability and colorectal cancer.
Grady WM, Markowitz S.
Curr. Opin. Gastroenterol. 2000 Jan;16(1):62-67.3.Carcinogenesis in the GI tract: from morphology to genetics and back again.
Redston M.
Mod. Pathol. 2001 Mar;14(3):236-45.
4.Wnt pathway mutations selected by optimal beta-catenin signaling for tumorigenesis.
Cho KH, Baek S, Sung MH.
FEBS Lett. 2006 Jun 26;580(15):3665-70.
5.Mutations in APC, Kirsten-ras, and p53–alternative genetic pathways to colorectal cancer.
Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR.
Proc. Natl. Acad. Sci. U S A. 2002 Jul 9;99(14):9433-8.
6.Adenomatous polyposis coli (APC)-independent regulation of beta-catenin degradation via a retinoid X receptor-mediated pathway.
Xiao JH, Ghosn C, Hinchman C, Forbes C, Wang J, Snider N, Cordrey A, Zhao Y, Chandraratna RA.
J. Biol. Chem. 2003 Aug 8;278(32):29954-62.
7.Prostaglandin E2 Stimulates the beta-catenin/T cell factor-dependent transcription in colon cancer.
Shao J, Jung C, Liu C, Sheng H.
J. Biol. Chem. 2005 Jul 15;280(28):26565-72.
8.Role of beta-arrestin 1 in the metastatic progression of colorectal cancer.
Buchanan FG, Gorden DL, Matta P, Shi Q, Matrisian LM, DuBois RN.
Proc. Natl. Acad. Sci. U S A. 2006 Jan 31;103(5):1492-7.
9.Signaling pathways in intestinal development and cancer.
Sancho E, Batlle E, Clevers H.
Annu. Rev. Cell Dev. Biol. 2004;20:695-723.
10.The role of MLH1, MSH2 and MSH6 in the development of multiple colorectal cancers.
Lawes DA, Pearson T, Sengupta S, Boulos PB.
Br. J. Cancer. 2005 Aug 22;93(4):472-7.
11.SMAD4 mutations in colorectal cancer probably occur before chromosomal instability, but after divergence of the microsatellite instability pathway.
Woodford-Richens KL, Rowan AJ, Gorman P, Halford S, Bicknell DC, Wasan HS, Roylance RR, Bodmer WF, Tomlinson IP.
Proc. Natl. Acad. Sci. U S A. 2001 Aug 14;98(17):9719-23.
12.Mediation of the DCC apoptotic signal by DIP13 alpha.
Liu J, Yao F, Wu R, Morgan M, Thorburn A, Finley RL Jr, Chen YQ.
J. Biol. Chem. 2002 Jul 19;277(29):26281-5.
13.Metastatic colorectal cancer.
Saletti P, Cavalli F.
Cancer Treat. Rev. 2006 Nov;32(7):557-71.
14.IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice.
Hanada T, Kobayashi T, Chinen T, Saeki K, Takaki H, Koga K, Minoda Y, Sanada T, Yoshioka T, Mimata H, Kato S, Yoshimura A.
J. Exp. Med. 2006 Jun 12;203(6):1391-7.
15.Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation.
Itzkowitz SH, Yio X.
Am. J. Physiol. Gastrointest. Liver Physiol. 2004 Jul;287(1):G7-17.
16.RNAi technology: a revolutionary tool for the colorectal cancer therapeutics.
Lv W, Zhang C, Hao J.
World J. Gastroenterol. 2006 Aug 7;12(29):4636-9.
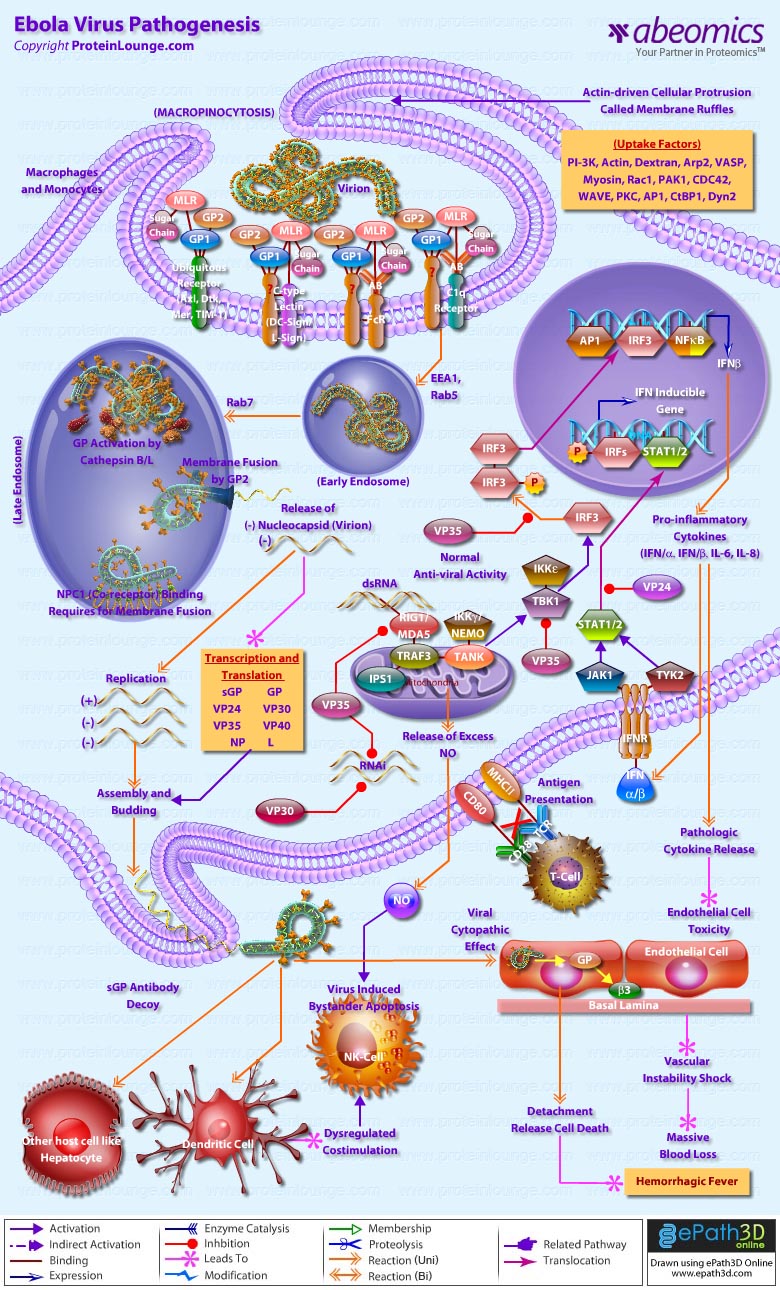
Ebola virus is an aggressive pathogen that causes a highly lethal hemorrhagic fever syndrome in humans and nonhuman primates. Typically, Ebola virus infection runs its course within 14 to 21 days. Infection initially presents with nonspecific flu-like symptoms such as fever, myalgia, and malaise. As the infection progresses, patients exhibit severe bleeding and coagulation abnormalities, including gastrointestinal bleeding, rash, and a range of hematological irregularities, such as lymphopenia and neutrophilia. Cytokines are released when reticulo-endothelial cells encounter virus, which can contribute to exaggerated inflammatory responses that are not protective. Damage to the liver, combined with massive viremia, leads to disseminated intravascular coagulopathy. The virus eventually infects microvascular endothelial cells and compromises vascular integrity. The terminal stages of Ebola virus infection usually include diffuse bleeding, and hypotensive shock accounts for many ebola virus fatalities (Ref.1).
Ebola virus and the related Marburg virus are members of the filovirus family, which are pleomorphic, negative-sense RNA viruses whose genome organization is most similar to the Paramyxoviridae. The Ebola virus genome is 19 kb long, with seven open reading frames encoding structural proteins, including the virion envelope glycoprotein (GP), nucleoprotein (NP), and matrix proteins VP24 and VP40; nonstructural proteins, including VP30 and VP35; and the viral polymerase. The GP open reading frame of Ebola virus gives rise to two gene products, a soluble 60- to 70-kDa dimeric soluble protein (sGP) and a full-length 150- to 170-kDa trimeric protein (GP) that inserts into the viral membrane, through transcriptional editing (Ref.2). This envelope GP spike is expressed at the cell surface, and is incorporated into the virion to drive viral attachment and membrane fusion. It has also been shown as the crucial factor for Ebola virus pathogenicity (Ref. 3). VP40 is located beneath the viral envelope where it helps to maintain structural integrity. It has also been associated with late endosomes and likely mediates filovirus budding due to its ability to induce its own release from cells in the absence of all other viral proteins (Ref.4). The second matrix protein, VP24, has been shown to suppress interferon production. However, interferon interference is not its only function. This protein, along with VP35 and NP are sufficient to form nucleocapsid structures (Ref.5). Lastly, VP24 is necessary for the correct assembly of a functional nucleocapsid. The remaining structural proteins form the nucleocapsid. These are the nucleoprotein NP, the polymerase cofactor VP35, the viral-specific transcription activator VP30 and the viral RNA polymerase L (Ref.2).
As the first step of the viral life cycle, entry into the host cell is a major and multi facet process. Although the mechanism is poorly understood, cellular entry involves uptake by a macropinocytosis-like mechanism. This mechanism is associated with outward extensions of the plasma membrane formed by actin polymerization. These so called membrane ruffles can fold back upon themselves and form a macropinosome upon fusion of the distal loop ends. Virus entry into cells is initiated by the interaction of the viral glycoprotein1 subunit (GP1) with both adherence factors and one or more receptors on the surface of host cells (Ref.6). GP is highly glycosylated with large amounts of N- and O-linked glycans, designated the mucin-like region (MLR). The MLR plays an important role in filovirus entry in to preferred target cells (Ref.7). A variety of C-type lectins (e.g., DC-SIGN, L-SIGN) expressed on key cell types (e.g.,macrophages, DCs) can mediate viral attachment and enhance viral infection (Ref.8). On epithelial cells, TIM-1 serves as a receptor for virus entry (Ref.6). Moreover, members of the Tyro3 receptor tyrosine kinase family (e.g., Axl, Dtk,and Mer) enhances GP-induced macropinocytic uptake (Ref.7).
Internalized virus bearing EBOV GP virions first co-localizes with EEA1 (early endosomal antigen-1)-positive compartments and are then trafficked to Rab5-positive early endosomes. Viral co-localization with perinuclear Rab7/LAMP-1-positive late endosomes can be observed at later times, and delivery to these compartments appears to be important for entry (Ref.8). Within the acidified endosome, the heavily glycosylated GP1 is cleaved to a smaller form by the low pH-dependent cellular proteases Cathepsin B and L, exposing RBS (residues in the receptor binding site), which is a ligand for NPC1 and mediates membrane fusion by the GP2 subunit. NCP1 is crucial for viral membrane fusion and escape from the lysosomal vesicle (Ref.6 & 7). Apart from the conventional micropinocytosis way of entry Filoviruses utilize another mechanism for entry known as antibody-dependent enhancement (ADE) of viral infection, where Filoviruses utilize virus-specific antibodies for their entry into cells in vitro through interaction between anti-GP antibodies and the cellular Fc receptor (FcR) or complement component C1q and its ligand, which likely promotes viral attachment to cells (Ref.7).The energetically unfavorable insertion of the EBOV GP2 fusion loop into host endosomal membranes is followed by the energetically favorable collapse of EBOV GP into a six-helix bundle, allowing for lipid mixing and hemifusion of host and viral membrane lipids. Finally, the hemifused host and viral membranes resolve and a complete pore is formed, through which the viral genomic complex passes into the cytoplasm, allowing the viral replication cycle to continue (Ref.6).
Once the viral and internal cell membranes fuse, the virus particle uncoats and its anti-genome is transcribed into mRNA using nucleocapsid-associated viral proteins. Transcription of the genome is mediated via a complex of VP30, VP35, and the viral polymerase L bound to an NP coated genome. VP30 phosphorylation leads to its dissociation from the VP35/L complex and is the signal to switch from transcription to replication. Following this switch, virus genomes are replicated and coated by NP, VP24, VP30, and VP35 During assembly, L associates with the ribonucleoprotein complex through an interaction with VP35. The ribonucleoproteins, then associate with the matrix protein VP40, and viral particles are extruded through the plasma membrane within lipid raft microdomain regions. Expression and secretion of sGP serves as an antibody decoy for antibodies generated against GP. Viral proteins VP35, VP30, and VP24 are expressed and mediate innate immune avoidance in all cell types. VP35 interferes with RIG-I/MDA-5 signaling and induction of Interferon. Additionally, VP35 and VP30 block the RNAi response against viral gene expression. VP24 acts to inhibit type I and II Interferon (IFN) signaling. This prevents Interferon-induced gene expression and, in antigen-presenting cells, blocks enhancement of antigen presentation to T cells (Ref.9).
During infection, monocytes /macrophages in the lymphoid tissues are early and sustained targets of this deadly virus. The white blood cells respond by releasing large amounts of proinflammatory cytokines that increase permeability of the vascular endothelium, which facilitates easier entry into the virus’s secondary targets, endothelial cells. Following GP-mediated macropinocytosis the cell detaches from its neighbors and loses contact with its basement membrane through a mechanism of glycan mediated steric occlusion by GP. The newly created particles then leave via lipid rafts, leaving a destabilized vascular system responsible for the massive blood loss causing hemorrhagic fever characteristic of Ebola patients. The pro-inflammatory cytokines also recruit more macrophages to the area, maximizing the number of cells that ebola can use to spread throughout the body. In the meantime, hepatocytes are being destroyed by the virus, ensuring that these cell signals cannot be cleared from the bloodstream. In addition, infected macrophages release increased amounts of nitric oxide (NO), a gaseous hormone that normally functions in cell communication. However, in high concentrations, NO depresses the mitochondrial membrane potential, causing apoptosis in nearby natural killer cells (Ref. 10 & 11).
1.Ebola virus pathogenesis: implications for vaccines and therapies.
Sullivan N, Yang ZY, Nabel GJ
J Virol. 2003 Sep;77(18):9733-7. Review. No abstract available.
2.Ebolavirus: a brief review of novel therapeutic targets.
Kondratowicz AS, Maury WJ.
Future Microbiol. 2012 Jan;7(1):1-4. doi: 10.2217/fmb.11.110. Review.
3.Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha.
Yonezawa A, Cavrois M, Greene WC.
J Virol. 2005 Jan;79(2):918-26.
4.Ebola virus VP40-induced particle formation and association with the lipid bilayer.
Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y.
J Virol. 2001 Jun;75(11):5205-14.
5.The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein.
Huang Y, Xu L, Sun Y, Nabel GJ.
Mol Cell. 2002 Aug;10(2):307-16.
6.Filovirus entry: a novelty in the viral fusion world.
Hunt CL, Lennemann NJ, Maury W.
Viruses. 2012 Feb;4(2):258-75. doi: 10.3390/v4020258. Epub 2012 Feb 7. Review.
7.Filovirus tropism: cellular molecules for viral entry.
Takada A.
Front Microbiol. 2012 Feb 6;3:34. doi: 10.3389/fmicb.2012.00034. eCollection 2012.
8.Filovirus entry into cells – new insights.
Miller EH, Chandran K.
Curr Opin Virol. 2012 Apr;2(2):206-14. doi: 10.1016/j.coviro.2012.02.015. Epub 2012 Mar 23. Review.
9.Camouflage and Misdirection: The Full-On Assault of Ebola Virus Disease.
Misasi J, Sullivan NJ.
Cell. 2014 Oct 23;159(3):477-486. doi: 10.1016/j.cell.2014.10.006. Epub 2014 Oct 16. Review.
10.Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection.
Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ.
Am J Pathol. 2003 Dec;163(6):2347-70.
11.Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein.
Francica JR, Varela-Rohena A, Medvec A, Plesa G, Riley JL, Bates P.PLoS Pathog. 2010 Sep 9;6(9):e1001098. doi: 10.1371/journal.ppat.1001098.
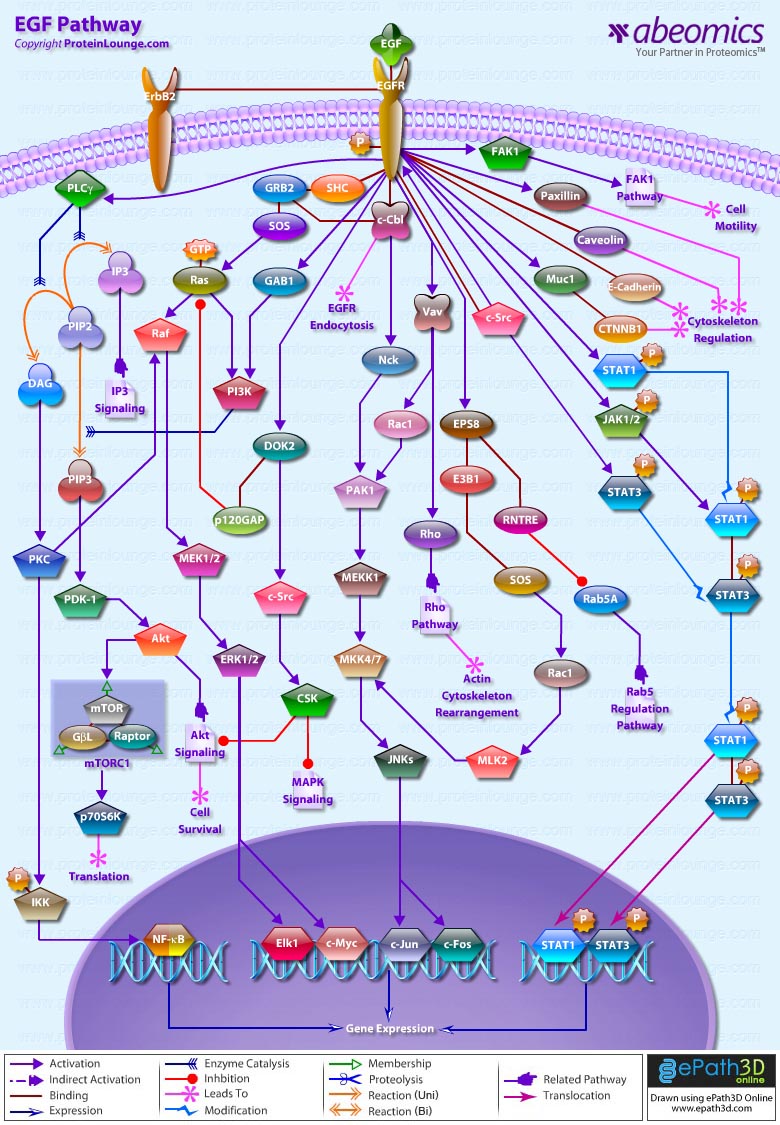
EGF (Epidermal Growth Factor) is a small 53 amino acid residue protein that is involved in normal cell growth, oncogenesis, and wound healing. This protein shows both strong sequential and functional homology with hTGF-Alpha (human type-Alpha Transforming Growth Factor), which is a competitor for EGF receptor sites. EGF binds to a specific high-affinity, low-capacity receptor on the surface of responsive cells known as EGFR (Epidermal growth factor receptor). EGFR is a member of the ErbB (Erythroblastic Leukemia Viral Oncogene Homolog) family receptors, a subfamily of four closely related receptor tyrosine kinases: EGFR (ErbB1), Her2/c-neu (ErbB2), Her3 (ErbB3) and Her4 (ErbB4). In response to toxic environmental stimuli, such as ultraviolet irradiation, or to receptor occupation by EGF, the EGFR forms Homo- or Heterodimers with other family members. Binding of EGF to the extracellular domain of EGFR leads to receptor dimerization, activation of the intrinsic PTK (Protein Tyrosine Kinase) activity, tyrosine autophosphorylation, and recruitment of various signaling proteins to these autophosphorylation sites located primarily in the C-terminal tail of the receptor. Tyrosine phosphorylation of the EGFR leads to the recruitment of diverse signaling proteins, including the Adaptor proteins GRB2 (Growth Factor Receptor-Bound Protein-2) and Nck (Nck Adaptor Protein), PLC-Gamma (Phospholipase-C-Gamma), SHC (Src Homology-2 Domain Containing Transforming Protein), STATs (Signal Transducer and Activator of Transcription), and several other proteins and molecules. The evolutionary conservation of all the components of the EGFR signaling pathway in Nematode, Fruit fly, Mouse, and Man underscores the biological significance of this signaling pathway. Furthermore, aberrant regulation of the activity or action of EGFR and other members of the RTK family have been implicated in multiple cancers, including those of brain, lung, mammary gland, and ovary (Ref.1 & 2).
GRB2 is an essential component of EGFR signaling to Ras. The SH2 (Src Homology-2) domain of GRB2 can bind directly to phosphotyrosines 1068 and 1086 of the activated EGFR or indirectly through the tyrosine-phosphorylated adaptor protein SHC. The SH3 domains of GRB2 are constitutively associated with SOS (Son of Sevenless), an exchange factor of Ras GTPase. Besides interaction with SOS, GRB2 SH3 domains are capable of association with several proteins, including Dynamin and Cbl (c-Cbl, Cbl-b, and Cbl-3), both implicated in the regulation of EGFR endocytosis. Binding of the GRB2 and SOS complex to the EGFR places SOS in proximity to Ras, thus leading to GTP-loading of Ras and subsequent activation of Ras effectors, such as Raf kinases and PI3K (Phosphatidylinositol 3-Kinase). Raf initiates a cascade of phosphorylation events including the phosphorylation and activation of the MEKs (MAPK/ERK Kinases) and ERKs (Extracellular Signal-Regulated Kinases). EGF stimulation of PI3K may also be mediated by the docking protein GAB1 (GRB2-Associated Binder-1). GAB1 is a docking protein that recruits PI3K and other effector proteins in response to the activation of many RTKs (Receptor Tyrosine Kinases). PI3K once activated, phosphorylates membrane bound PIP2 (Phosphatidylinositol (4,5)-bisphosphate) to generate PIP3 (Phosphatidylinositol-3,4,5-trisphosphate). The binding of PIP3 to the PH domain anchors Akt to the plasma membrane and allows its phosphorylation and activation by PDK1 (Phosphoinositide-Dependent Kinase-1). Akt then phosphorylates several substrates and take part in cell survival (Ref.3 & 4). One of the prominent enzymes activated by EGFR is the G1 isoform of PLC (Phospholipase-C-Gamma1). This enzyme, which has two SH2 domains, catalyses the hydrolysis of PIP2, generating the second messengers DAG (1,2-Diacylglycerol) and IP3 (Inositol Trisphosphate). IP3 diffuses through the cytosol and releases stored Ca2+ (Calcium) ions from the ER (Endoplasmic Reticulum). DAG is the physiological activator of PKC (Protein Kinase-C), which in turn lead to phosphorylation of various substrate proteins that are involved in an array of cellular events. PKC also leads to the activation of IKKs (I-KappaB-Kinases), and finally nuclear factor NF-KappaB (Nuclear Factor-KappaB)-dependent transcription (Ref.5).
Dok2 (Docking Protein-2) also associates with the EGFR and become tyrosine phosphorylated in response to EGF stimulation. The recruitment of Dok2 to the EGFR, which is mediated through its PTB (Phosphotyrosine Binding) domain, results in attenuation of MAPK (Mitogen-Activated Protein Kinase) activation. Dok2’s ability to attenuate EGF-driven MAPK activation is independent of its ability to recruit RasGAP, a known attenuator of MAPK activity, suggesting an alternate Dok2-mediated pathway. Dok2 associate with c-Src and with the SFK (Src Family Kinase)-inhibitory kinase, Csk. Dok2 associates constitutively with c-Src through an SH3-dependent interaction and that this association is essential to Dok2’s ability to attenuate c-Src activity and diminish MAPK and Akt/PKB activity (Ref.6). One of the important signaling events activated by EGFR involves tyrosine phosphorylation of STAT. Stimulation of EGFR induces Tyrosine phosphorylation of STAT1 and STAT3 and initiates complex formation of STAT1 and STAT3 with JAK1 (Janus Kinase-1) and JAK2 (Janus Kinase-2). JAKs are essential to mediate interaction of EGFR with STAT1 and STAT3. Thereafter, the STATs translocate to the nucleus, where they are active in gene transcription. EGFR also activate STAT3 in a manner largely independent of JAKs but dependent upon the activation of Src kinases. c-Src is activated by EGF-induced EGF receptor activation. Activated c-Src phosphorylates EGF receptor on tyrosine 845 that plays an important role in tyrosine phosphorylation and activation of STAT proteins (Ref.2 & 7).
E3B1 (EPS8 Binding Protein)/ABI1 (Abl-Interactor-1) is a protein involved in the EGFR signalling pathway. E3B1/ABI1 interacts with EPS8 (Epidermal Growth Factor Receptor Pathway Substrate-8), a protein which is phosphorylated in fibroblasts in response to EGF. Remarkably, E3B1/ABI1 is a negative regulator of the cellular response mediated by growth factor receptors. EPS8 binds, through its SH3 domain, to either E3B1 or RNTRE (Related to the N-terminus of tre). EPS8 mediates the transfer of signals between Ras and Rac, by forming a complex with E3B1 and SOS1. SOS1, a bifunctional GEF (Guanine nucleotide Exchange Factor), activates Ras in vivo and displays Rac-GEF activity in vitro, when engaged in a tricomplex with EPS8 and E3B1. On the other hand, by entering in a complex with EPS8, RNTRE acts on Rab5A (Rab5A, member RAS oncogene family) and inhibits internalization of the EGFR (Ref.8). Cbl is another major substrate for the EGFR that associates with this receptor upon EGF stimulation, and forms complexes with several signaling proteins that play key roles in EGF-mediated cell growth. Since Nck is known to bind to activated EGFR through its SH2 domain and to Cbl via its SH3 domains, Nck represented another potential adaptor that could mediate Cbl-EGFR association. The SH3-SH3-SH3-SH2 adapter protein Nck links EGFR to downstream signaling pathways, among which p21CDC42 (Cell Division Cycle-42) /Rac-activated kinase cascade, SOS-activated Ras signaling and the human WASP (Wiskott-Aldrich Syndrome protein)-mediated actin cytoskeleton changes, have been implicated. Nck also activates PAK1 (p21/CDC42/Rac1-Activated Kinase-1). The association between Nck and PAK1 has been shown to occur through the first N-terminal polyproline domain of PAK1 and an SH3 domain of Nck. PAK activated by Nck activates JNKs (c-Jun Kinases) via MEKK1 (MAP/ERK Kinase Kinase-1) and MKK4/7 (MAP Kinase Kinase-4/7) respectively. JNKs once activated enter the nucleus and causes phosphorylation of transcription factors such as c-Fos and c-Jun. EGFR also activates Vav (Oncogene Vav). Vav proteins are guanine nucleotide exchange factors for Rho family GTPases which activate pathways leading to actin cytoskeletal rearrangements and transcriptional alterations. Each Vav protein co precipitated with activated EGF and multiple phosphorylated tyrosine residues on the EGFR were able to mediate Vav2 tyrosine phosphorylation. Vav activates Rho or Rac. The Rho family GTPase Rac initiates a cascade leading to JNK/SAPK, presumably by binding and activating the protein kinase, a kinase that phosphorylates and promotes activation of MEKK1. Rho, activated by Vav, is responsible for actin cytoskeletal rearrangement (Ref.9 & 10).
EGFR also activates several other proteins including FAK (Focal Adhesion Kinase), Paxillin, Caveolin, E-Cadherin and Ctnn-Beta (Catenin-Beta). FAK1 take part in cell motility, whereas Caveolin, Cadherin and Ctnn-beta are involved ibn Cytoskeletal Regulation. Activation of Ctnn-Beta occurs via Muc1 (Mucin-1). Activated EGFR phosphorylates the Muc1 cytoplasmic tail on tyrosine at a YEKV motif that functions as a binding site for the c-Src SH2 domain. EGFR also increases binding of Muc1 and Ctnn-Beta. Thus, EGFR regulates interactions of Muc1 with c-Src and Ctnn-beta. EGFR is overexpressed or activated by autocrine growth factors in many types of tumors, including breast, thyroid, ovarian, colon, head and neck, and brain. Furthermore, EGFR overexpression has been linked to a poor prognosis in breast cancer and may promote proliferation, migration, invasion, and cell survival as well as inhibition of apoptosis. EGFR-targeted therapies offer the promise of better treatment for many types of solid tumors, including non-small cell lung cancer. Anti-EGFR agents include mAbs (Monoclonal Antibodies) targeting the EGFR extracellular receptor domain and small-molecule TKIs (Tyrosine Kinase Iinhibitors) targeting the EGFR intracellular kinase domain. Both mAbs and TKIs have demonstrated encouraging results as monotherapies and in combination with chemotherapy and radiotherapy (Ref.11 & 12).
1.EGFR-expression in pulmonary neuroendocrine cell hyperplasia.
Kuhnen C, Winter BU.
Pathologe. 2006 Mar;27(2):147-51.
2.Multiple signaling pathways mediate compaction of collagen matrices by EGF-stimulated fibroblasts.
Smith KD, Wells A, Lauffenburger DA.
Exp Cell Res. 2006 Apr 2;
3.Molecular analysis of the EGFR-RAS-RAF pathway in pancreatic ductal adenocarcinomas: lack of mutations in the BRAF and EGFR genes.
Immervoll H, Hoem D, Kugarajh K, Steine SJ, Molven A.
Virchows Arch. 2006 Apr 6;
4.Participation of both Gab1 and Gab2 in the activation of the ERK/MAPK pathway by epidermal growth factor.
Meng S, Chen Z, Munoz-Antonia T, Wu J.
Biochem J. 2005 Oct 1;391(Pt 1):143-51.
5.Akt Binds to and Phosphorylates Phospholipase C-{gamma}1 in Response to Epidermal Growth Factor.
Wang Y, Wu J, Wang Z.
Mol Biol Cell. 2006 May;17(5):2267-77. Epub 2006 Mar 8.
6.Dok-R mediates attenuation of epidermal growth factor-dependent mitogen-activated protein kinase and Akt activation through processive recruitment of c-Src and Csk.
Van Slyke P, Coll ML, Master Z, Kim H, Filmus J, Dumont DJ.
Mol Cell Biol. 2005 May;25(9):3831-41.
7.Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor.
Alvarez JV, Greulich H, Sellers WR, Meyerson M, Frank DA.
Cancer Res. 2006 Mar 15;66(6):3162-8.
8.The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5.
Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, Di Fiore PP.
Nature. 2000 Nov 16;408(6810):374-7.
9.Sprouty2 acts at the Cbl/CIN85 interface to inhibit epidermal growth factor receptor downregulation.
Haglund K, Schmidt MH, Wong ES, Guy GR, Dikic I.
EMBO Rep. 2005 Jul;6(7):635-41.
10.Role of group A p21-activated kinases in activation of extracellular-regulated kinase by growth factors.
Beeser A, Jaffer ZM, Hofmann C, Chernoff J.
J Biol Chem. 2005 Nov 4;280(44):36609-15. Epub 2005 Aug 29.
11.The epidermal growth factor receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and beta-catenin.
Li Y, Ren J, Yu W, Li Q, Kuwahara H, Yin L, Carraway KL 3rd, Kufe D.
J Biol Chem. 2001 Sep 21;276(38):35239-42. Epub 2001 Aug 1.
12.Clinical Implications of EGFR Expression in the Development and Progression of Solid Tumors: Focus on Non-Small Cell Lung Cancer.
Ettinger DS.Oncologist. 2006 Apr;11(4):358-73.
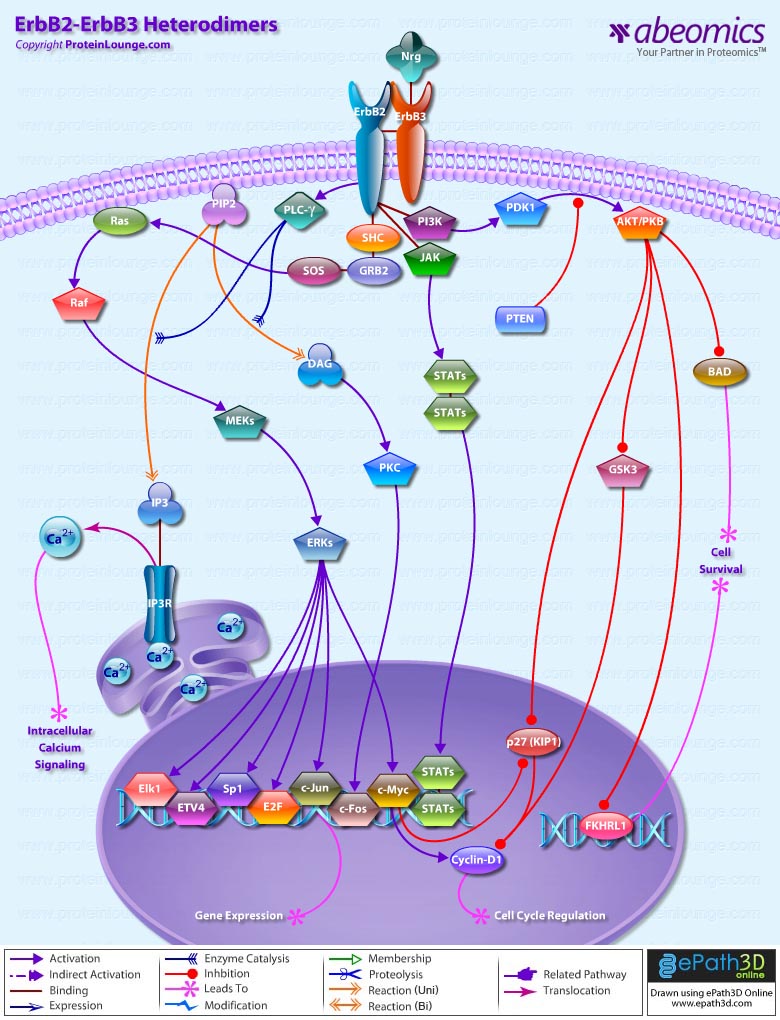
The ErbB (Erythroblastic Leukemia Viral Oncogene Homolog) family of transmembrane RTKs (Receptor Tyrosine Kinases) plays an important role in the pathogenesis of many cancers (Ref.1). This family is comprised of four members EGFR (Epidermal Growth Factor Receptor), ErbB2, ErbB3, and ErbB4 (Ref.2). ErbB2 also called Her2 (Heregulin-2) and ErbB3 are closely related to the EGFR/ErbB1, but unlike EGFR, ErbB2 is a ligandless receptor, whereas ErbB3 lacks tyrosine kinase activity. Hence, both ErbB2 and ErbB3 are active only in the context of ErbB heterodimers, and ErbB2-ErbB3 heterodimers, which are driven by Nrg (Neuregulin) ligands, are the most prevalent and potent complexes in terms of cell growth and transformation. The basis for the potency of signaling by the ligand-activated ErbB2-ErbB3 heterodimer lies in the fact that this dimer has the capacity to signal very potently, both through the Ras-ERK (Extracellular Signal Regulated Kinase) pathway for proliferation, and through the PI3K-Akt/PKB (Phosphatidylinositol-3 Kinase-Akt/Protein Kinase-B) pathway for survival. In addition, this receptor dimer evades downregulation mechanisms, leading to prolonged signaling.
Ligand-induced formation of the ErbB2-ErbB3 heterodimer at the cell surface leads to activation of several major pathways of signal transduction. This process results in enhanced cell survival and mitogenicity, and its deregulation can lead to tumorigenesis. Stimulation of ERK occurs upon ligand-induced activation of a receptor dimer, which binds GRB2 (Growth Factor Receptor-Bound Protein-2)through a phosphorylated tyrosine-based consensus site, or indirectly, through interaction with SHC. GRB2 is associated with SOS (Son of Sevenless), a guanine nucleotide exchange factor specific for Ras, and SOS activates Ras by exchanging GDP for GTP. In the GTPase active state, Ras interacts with Raf and stimulates a linear kinase cascade culminating in activation of ERK/MAPK (Mitogen-Activated Protein Kinases). ERK phosphorylates a variety of cytoplasmic and membranal substrates, and is rapidly translocated to the nucleus, where it activates a number of transcription factors including Sp1, PEA3, E2F, Elk1, Jun, Fos and Myc>c-Myc oncoprotein, which is a major transcription factor and regulator of cell cycle progression.
Another pathway is the P13K-Akt pathway. Activation of PI3K occurs through binding of the regulatory p85 subunit of the lipid kinase to a Phosphotyrosine consensus site on the receptor, leading to allosteric activation of the p110 catalytic subunit. p110 activation produces PIP3 (Phosphatidylinositol-3,4,5-Trisphosphate) from PIP2 (Phosphatidylinositol 4,5-Bisphosphate). The PH domain-containing proteins PDK1 (Phosphoinositide-Dependent Protein Kinase-1) and Akt/PKB are key mediators of PI3K signaling, and both are essential for the transforming effects of PI3K. Upon production of PIP2 and PIP3 following activation of PI3K by the ErbB2-ErbB3 receptor dimer, Akt is recruited to the plasma membrane by its PH domain, and is phosphorylated by PDK1. Akt phosphorylation causes its activation and translocation to the nucleus, where it acts upon its targets, which are either regulators of apoptosis or of cell growth, through inhibition of the proapoptotic proteins BAD (BCL2 Antagonist of Cell Death), GSK3 (Glycogen Synthase Kinase-3), and the transcription factor FKHR-L1. The tumor suppressor PTEN (Phosphatase and Tensin Homolog Deleted On Chromosome 10) is a lipid phosphatase, which dephosphorylates the 3’-OH position of PIP2 and PIP3, thereby reverting the activity of PI3K, and downregulating the activity of PDK1 and Akt. In addition, the PLC-Gamma (Phospholipase-C-Gamma) and the JAK/STAT (Janus Kinase/ Signal Transducers and Activators of Transcription Factor) pathways are indicated, with their resulting enhancement of transcription leading to cell proliferation. Activation of PLC-Gamma occurs through its SH2-mediated recruitment to phosphorylation- dependent docking sites on ErbB2, as well as recruitment through its PH domain to the plasma membrane. In its phosphorylated active form, PLC-Gamma hydrolyzes PIP2 into IP3 (Inositol Triphosphate), and DAG (Diacylglycerol). IP3 activates the release of Ca+2 (Calcium) from intracellular stores, and thereby activates Ca+2/Calm (Calmodulin) dependent kinases, as well as additional pathways, and it collaborates with DAG to stimulate PKC (Protein Kinase-C).
A major player acting downstream of ErbB2-ErbB3 is Cyclin-D1. A number of pathways lead from the receptors to enhanced activation of Cyclin-D1, thereby promoting cell cycle progression. The outcome of activation of these different signaling pathways depends on the cellular context, and can vary from proliferation to differentiation, migration, and even induction of apoptosis (Ref.3).
Nagy P, Vereb G, Sebestyen Z, Horvath G, Lockett SJ, Damjanovich S, Park JW, Jovin TM, Szollosi J.
J Cell Sci. 2002 Nov 15;115(Pt 22):4251-62.2.ErbB2 is required for muscle spindle and myoblast cell survival.
Andrechek ER, Hardy WR, Girgis-Gabardo AA, Perry RL, Butler R, Graham FL, Kahn RC, Rudnicki MA, Muller WJ.
Mol Cell Biol. 2002 Jul;22(13):4714-22.3.The deaf and the dumb: the biology of ErbB-2 and ErbB-3.
Citri A, Skaria KB, Yarden Y.
Exp Cell Res. 2003 Mar 10;284(1):54-65.
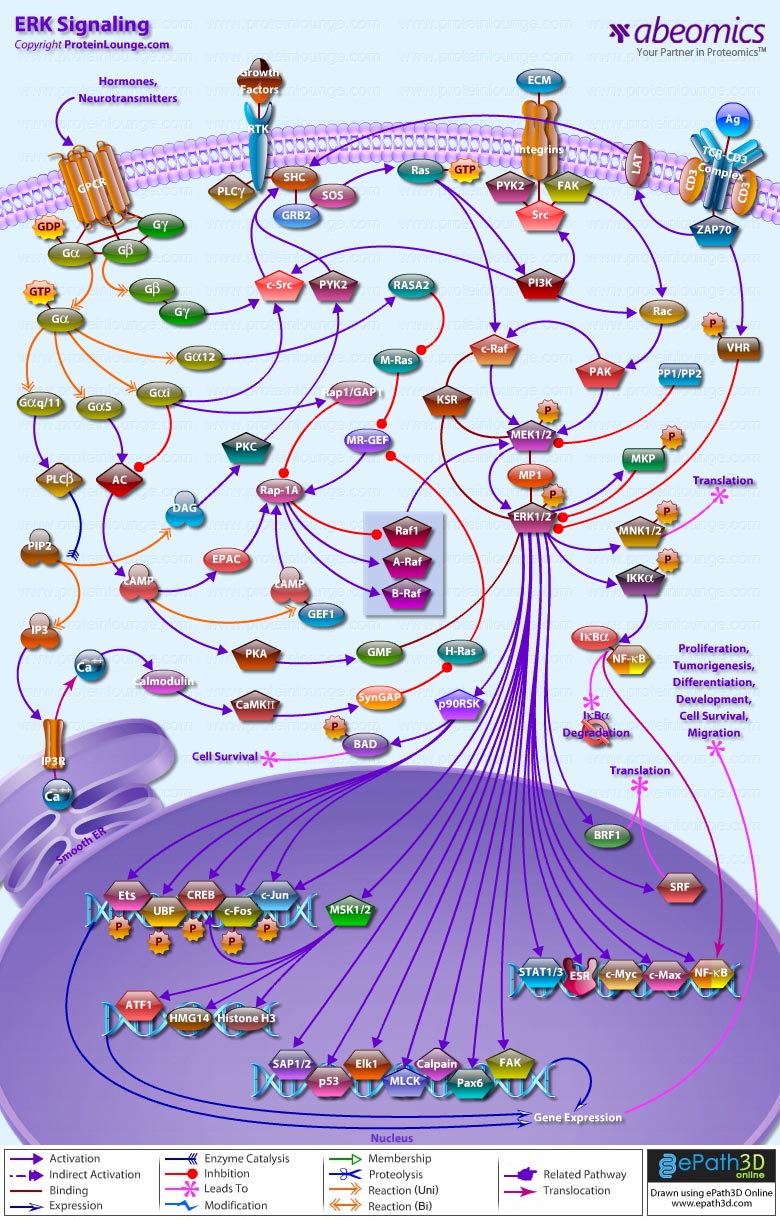
The MAPK (Mitogen-Activated Protein Kinase) pathway is one of the primordial signaling systems that nature has used in several permutations to accomplish an amazing variety of tasks. It exists in all eukaryotes, and controls such fundamental cellular processes as Proliferation, Differentiation, Survival and Apoptosis. Mammalian MAPK can be divided into four groups based on their structure and function: ERKs (Extracellular signal-Regulated Kinases), p38MAPKs, JNKs (c-Jun NH2-terminal Kinases) and ERK5 (Extracellular signal-Regulated Kinase-5) or BMK. Activation of these MAPKs occurs through a cascade of upstream kinases; a MAPKKK (MAPK Kinase Kinase) first phosphorylates a dual-specificity protein kinase MAPKK (MAPK Kinase), which in turn phosphorylates the MAPK. This set-up provides not only for signal amplification, but, maybe even more importantly, for additional regulatory interfaces that allow the kinetics, duration and amplitude of the activity to be precisely tuned. ERK, a member of the MAPK family, have been established as major participants in the regulation of cell growth and differentiation, but when improperly activated contribute to malignant transformation. ERK1 and 2 form a central component in the MAPK cascade. The MAPK/ERK signaling cascade is activated by a wide variety of receptors involved in growth and differentiation including GPCRs (G-Protein Coupled Receptors), RTKs (Receptor Tyrosine Kinases), Integrins, and Ion channels. The specific components of the cascade vary greatly among different stimuli, but the architecture of the pathway usually includes a set of adaptors like SHC, GRB2 (Growth Factor Receptor Bound protein-2), Crk, etc. linking the receptor to a GEF (Guanine nucleotide Exchange Factor) like SOS (Son of Sevenless), C3G, etc. transducing the signal to small GTP binding proteins (Ras, Rap1), which in turn activate the core unit of the cascade composed of a MAPKKK (Raf), a MAPKK (MEK1/2 (MAPK/ERK Kinase-1/2)) and MAPK (ERK). An activated ERK dimer can regulate targets in the cytosol and also translocate to the nucleus where it phosphorylates a variety of transcription factors regulating gene expression (Ref.1 & 2).
GPCRs constitute a superfamily of Plasma membrane receptors. Members of this family include receptors for many Hormones, Neurotransmitters, Chemokines and Calcium ion, as well as sensory receptors for various odors, and bitter and sweet tastes. GPCR play an important role in activation of ERKs. Stimulation of the GN-AlphaI (Guanine Nucleotide Binding Protein-Alpha Inhibiting Activity Polypeptide)-coupled Neuropeptide Y1 and GN-AlphaQ (Guanine Nucleotide-Binding Protein-Alpha-Q) -coupled Muscarinic M1 Acetylcholine Receptors lead to the activation of ERK. When the GPCR becomes activated, it leads to the exchange of GDP for GTP on the GN-Alpha subunit. Upon activation, GN-AlphaI or GN-AlphaQ subunits are separated from GN-Beta (Guanine Nucleotide-Binding Protein-Beta) and GN-Gamma (Guanine Nucleotide-Binding Protein-Gamma) subunits and are converted to their GTP bound states that exhibit distinctive regulatory features on the nine tmACs (Transmembrane Adenylate Cyclases) in order to regulate intracellular cAMP (Cyclic Adenosine 3′,5′-monophosphate) levels. cAMP activate Rap1A (Ras-Related Protein-1A) and Rap1B (Ras-Related Protein Rap1B) through EPAC (Exchange Protein Activated by cAMP)-dependent pathway. cAMP activates cAMP-GEFI (cAMP-Regulated Guanine Nucleotide Exchange Factor-I)/EPAC1 and cAMP-GEFII (cAMP-Regulated Guanine Nucleotide Exchange Factor-II)/EPAC2 that in turn activate Rap1A and Rap1B, respectively. Rap1A and Rap1B then forms an active complex with BRaf (v-Raf Murine Sarcoma Viral Oncogene Homolog-B1) for MEK1/2 activation finally resulting in ERK1/2 activation. cAMP may also activate PKA (Protein Kinase-A), which may further activate Rap and thus BRaf. On the other hand, PKA also inactivates C-Raf. GN-Alpha also directly activate PI3K (Phosphatidylinositide-3 Kinase) and c-Src and modulate their activity through stimulation of Ras under the influence of many extracellular factors. Rac is also a key downstream target/effector of PI3K. A new mechanism have been identified that regulates MEK1-ERK interactions and is dependent on Rac and PAK (p21-Activated Kinase). Besides GN-Alpha subunit, GN-Beta Gamma complex can also lead to activation of ERK. GN-Beta and GN-Gamma subunits when activated separate from GN-alpha subunit and it further activates PKC (Protein Kinase-C) via PLC-Beta (Phospholipase-C-Beta). PLC-Beta converts PIP2 (Phosphatidylinositol 4,5-bisphosphate) to DAG (Diacylglycerol), which activates PKC. PKC once formed can activate ERKs via Ras, Raf and MEKs. PKC also activates Src-PYK2 (Proline-Rich Tyrosine Kinase-2) complex which activates GRB2, which is also involved in ERK activation (Ref.3 & 4).
The canonical ERK MAPK cascade is also stimulated upon the binding of extracellular Growth factors to their respective transmembrane RTKs (Receptor Tyrosine Kinases). The subsequent auto-phosphorylation of the cytoplasmic tails of the receptor on tyrosine leads to the tyrosine phosphorylation of the adapter protein SHC. SHC can then recruit the GRB2-SOS complex to the membrane via the SH2 domain of GRB2 binding to the phosphotyrosine on SHC. SOS, a GEF for Ras, can then exchange the GDP bound to Ras to GTP. Once Ras binds GTP, it can then recruit the serine/threonine kinase Raf to the membrane. When Raf translocate to the membrane, it becomes activated and then phosphorylates the dual specificity kinase MEK. MEK binds and restricts inactive ERK to the cytosol. The MEK and ERK complex dissociates when MEK is activated and phosphorylates ERK. The ERK may then dimerize and this dimerization is apparently required for ERK to translocate into the nucleus by an active functions. Growth Factors may also activate ERK through PLC-Gamma and PKC. Integrins also play an important role in regulating the efficiency of the RTK/Ras/ERK pathway. Integrin regulation occurs at three (or more) different loci within the ERK pathway. First, in some cell systems, Integrin engagement with ligands enhances the activation and autophosphorylation of the RTKs. Secondly, Integrin engagement enhances the efficiency of the cytoplasmic cascade comprising Raf1, MEK and ERK. Finally, Integrin engagement is necessary for trafficking of activated ERK from the cytoplasm to the nucleus. FAK (Focal Adhesion Kinase) is a major nonreceptor tyrosine kinase activated after Integrin-mediated adhesion to ECM (Extracellular Matrix) proteins such as FN (Fibronectin). Interaction between FAK and the cytoplasmic tail of Beta1 Integrins results in autophosphorylation of FAK tyrosine 397 (FAK pY397) that can lead to stimulation of a cell-signaling cascade that ultimately activates the Ras/MAPK/ERK pathway. In addition to FAK, members of the Src family of nonreceptor protein-tyrosine kinases also associate with focal adhesions and are involved in Integrin signaling. Interestingly, Src and FAK appear to function in association with each other as a result of the binding of the Src SH2 domain to an autophosphorylation site of FAK. Src then phosphorylates additional sites on FAK. Tyrosine phosphorylation of FAK creates binding sites for the SH2 domains of other downstream signaling molecules, including PI3K and Rac. A key target of Rac is the protein-serine/threonine kinase PAK. Rac and CDC42 (Cell Division Cycle-42) can synergize with Raf to promote activation of the ERKs through mechanisms involving PAK1 phosphorylation of the MEK1 proline-rich sequence and PAK3 phosphorylation of Raf1. PAK3 can phosphorylate Raf1, enhancing Raf1 activation. Raf1 finally activates ERK1/2 via MEK1/2. The non-receptor tyrosine kinase PYK2 appears to function at a point of convergence of Integrins and certain GPCR signaling cascades. PYK2 acts with Src to link GN-AlphaI- and GN-AlphaQ-coupled receptors with GRB2 and SOS to activate the ERK signalling pathway (Ref. 5 & 6).
TCR (T-Cell Receptor)-CD3 complex also plays an important role in regulating ERK pathways in T-Cells. VHR is particularly interesting from a TCR signalling point of view because it accumulates at the T-Cell/APC (Antigen-Presenting-Cell) contact site, where it is phosphorylated at Tyr-138 by ZAP70 (Zeta-Chain-Associated Protein Kinase). This phosphorylation is required for VHR to inhibit the ERK MAPKs, giving ZAP70 an unanticipated control over MAPK dephosphorylation, in addition to its role as upstream activator of the Ras/Raf/MEK pathway. Other negative regulators of ERK pathway include PP2A (Protein Phosphatase-2A) and MKPs (MAPK Phosphatase). PP2A inhibits ERK pathway by dephosphorylating MEKs and is involved in the control of many cellular functions including metabolism, transcription, translation, RNA splicing, DNA replication, cell cycle progression, transformation, and apoptosis. The mammalian MAPK signaling system employ scaffold proteins, in part, to organize the MAPK signaling components into functional MAPK modules, thereby enabling the efficient activation of specific MAPK pathways. The ERK scaffold protein KSR (kinase suppressor of Ras) binds ERK, its direct activator MEK and Raf. In unstimulated cells KSR is mainly cytoplasmic but translocates to the Plasma membrane after growth factor stimulation, thus targeting MEK and ERK to the plasma membrane. A second targeting protein, p14, targets ERK2 to an endosomal location through its interaction with MP1 (MAPKK1-Interacting Protein-1), an adaptor protein that binds MEK and ERK. In addition, MEKK1 (MAP/ERK Kinase Kinase-1) can serve both as a scaffold and as MAPKKK, interacting specifically with MAPKK and MAPK (Ref.5, 7 & 8).
ERK once activated can either translocate to the nucleus to phosphorylate and activate transcription factors, while other pools of activated ERK phosphorylate a number of cytoplasmic targets. Cytosolic substrates for ERK include several pathway components involved in ERK negative feedback regulation. Multiple residues on SOS are phosphorylated by ERK following growth factor stimulation. SOS phosphorylation destabilizes the SOS-GRB2 complex, eliminating SOS recruitment to the plasma membrane and interfering with Ras activation of the ERK pathway. Negative feedback by ERK also occurs through direct phosphorylation of the EGF (Epidermal Growth Factor) receptor at Thr669. Finally, ERKs have also been demonstrated to negatively regulate themselves by phosphorylating MKPs (MAP Kinase Phosphatases) , which reduces the degradation of these phosphatases through the Ubiquitin-directed Proteasome complex. ERK also activate MNKs (MAPK-Interacting Kinases) by phosphorylation at Thr197 and Thr202. MNKs upregulate eIF4E (eukaryotic initiation factor-4E) through phosphorylation at Ser209 and play an important role in translation or they may also phosphorylate PLA2 (Phospholipase-A2). ERK1 and ERK2 regulate transcription indirectly by phosphorylating the RSKs (90 kDa Ribosomal protein S6 Kinases). Active RSKs appear to play a major role in transcriptional regulation, translocating to the nucleus and phosphorylating such factors as the product of proto-oncogene c-Fos at Ser362, SRF (Serum Response Factor) at Ser103, and CREB (Cyclic AMP Response Element-Binding protein) at Ser133. p90RSK also play an important role in cell survival by phosphorylating BAD (Bcl2-Antagonist of Cell Death). Another important cytoplasmic target of ERK is IKK-Alpha (I-KappaB Kinase-Alpha). IKK-Alpha phosphorylates I-KappaB-Alpha, which leads to ubiquitination and then leads to the degradation of I-KappaB-Alpha by the Proteosome, resulting in the translocation of NF-KappaB to the nucleus. In the nucleus it binds to its consensus sequence (5′-GGGACTTTC-3′) and positively regulates the transcription of genes involved in immune and inflammatory responses, cell growth control, and apoptosis (Ref.9, 10 & 11).
Upon phosphorylation, nuclear translocation of ERK1 and ERK2 is critical for both gene expression and DNA replication induced by growth factors. In the nucleus, ERK phosphorylates an array of targets, including transcription factors and a family of RSK-related kinases, the MSKs (Mitogen- and Stress-activated protein Kinases). MSKs phosphorylate and activate the AP1 component ATF1 (Activating Transcription Factor-1) at Ser63, and may be more important in vivo than RSKs in CREB phosphorylation at the activating Ser133. MSKs were also found to phosphorylate Histone H3 at Ser10 and Ser28, and the HMG14 (High-Mobility-Group protein-14) at Ser6, facilitating the rapid induction of immediate early genes following mitogenic stimulation. Probably the best-characterized transcription factor substrates of ERKs are TCFs (Ternary Complex Factors), including Elk1, which is directly phosphorylated by ERK1 and ERK2 at multiple sites, including the activating Ser383. Upon complex formation with SRF, phosphorylated TCFs transcriptionally activate the numerous Mitogen-inducible genes regulated by SREs (Serum Response Elements). TCFs Sap1 and Sap2 are also phosphorylated by ERK, as are other Ets family members. Another direct target of ERK is the product of proto-oncogene c-Myc, a short-lived transcription factor involved in multiple aspects of growth control. Following phosphorylation at Thr58 and Ser62 within its transactivation domain, Myc activates transcription as a heterodimeric partner with Max. ERK can also phosphorylate CREB directly as well as AP1 components c-Jun and c-Fos. ERK1/2 also phosphorylates MLCK (Myosin Light Polypeptide Kinase), Capn (Calpain), Pax6 (Paxillin-6) and FAK that play important role in Cytoskeletal rearrangement. Other ERK targets include STAT1/3 (Signal Transducer and Activator of Transcription-1/3) and ESR (Estrogen Receptor). Finally, it can be concluded that ERKs are involved in the regulation of important neuronal functions, including neuronal plasticity in normal and pathological conditions. The kinetics and localization of ERK are intrinsically linked, in that the duration of ERK activation dictates its subcellular compartmentalization and/or trafficking. The latter, in turn, dictates whether ERK-expressing cells would enter a program of cell death, survival or differentiation. With aberrations in the ERK cascade implicated in a high proportion of human cancers, many emerging therapies target proteins in the pathway. As these candidate therapies against ERK signaling components undergo development and enter trials, reagents that monitor their targets’ inhibition are critical for future success (Ref.12, 13 & 14).
Wada T, Penninger JM.
Oncogene. 2004 Apr 12;23(16):2838-49.2.Extracellular signal-regulated kinase as an inducer of non-apoptotic neuronal death.
Subramaniam S, Unsicker K.
Neuroscience. 2006;138(4):1055-65. Epub 2006 Jan 25.3.GPCR screening via ERK 1/2: a novel platform for screening G protein-coupled receptors.
Osmond RI, Sheehan A, Borowicz R, Barnett E, Harvey G, Turner C, Brown A, Crouch MF, Dyer AR.
J Biomol Screen. 2005 Oct;10(7):730-7. Epub 2005 Aug 29.
4.PKC{delta} mediates activation of ERK1/2 and induction of iNOS by Interleukin-1{beta} in Vascular Smooth Muscle Cells.
Ginnan R, Guikema BJ, Singer HA, Jourd’heuil D.
Am J Physiol Cell Physiol. 2006 Jan 25;
5.An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival.
Yang W, Klaman LD, Chen B, Araki T, Harada H, Thomas SM, George EL, Neel BG.
Dev Cell. 2006 Mar;10(3):317-27.
6.Adhesion stimulates direct PAK1/ERK2 association and leads to ERK-dependent PAK1 Thr212 phosphorylation.
Sundberg-Smith LJ, Doherty JT, Mack CP, Taylor JM.
J Biol Chem. 2005 Jan 21;280(3):2055-64. Epub 2004 Nov 12.
7.A novel PKC regulates ERK activation and degranulation of cytotoxic T lymphocytes: Plasticity in PKC regulation of ERK.
Puente LG, He JS, Ostergaard HL.
Eur J Immunol. 2006 Mar 21;36(4):1009-1018
8.The molecular scaffold kinase suppressor of Ras 1 (KSR1) regulates adipogenesis.
Kortum RL, Costanzo DL, Haferbier J, Schreiner SJ, Razidlo GL, Wu MH, Volle DJ, Mori T, Sakaue H, Chaika NV, Chaika OV, Lewis RE.
Mol Cell Biol. 2005 Sep;25(17):7592-604.
9.Protein-protein interactions in the regulation of the extracellular signal-regulated kinase.
Chuderland D, Seger R.
Mol Biotechnol. 2005 Jan;29(1):57-74.
10.Features of the catalytic domains and C termini of the MAPK signal-integrating kinases Mnk1 and Mnk2 determine their differing activities and regulatory properties.
Parra JL, Buxade M, Proud CG.
J Biol Chem. 2005 Nov 11;280(45):37623-33. Epub 2005 Sep 14.
11.p38 MAPK regulates phosphorylation of Bad via PP2A-dependent suppression of the MEK1/2-ERK1/2 survival pathway in TNF-alpha induced endothelial apoptosis.
Grethe S, Porn-Ares MI.
Cell Signal. 2006 Apr;18(4):531-40. Epub 2005 Jun 21.
12.Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2.
Knauf U, Tschopp C, Gram H.
Mol Cell Biol. 2001 Aug;21(16):5500-11.
13.Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism.
Butcher GQ, Lee B, Cheng HY, Obrietan K.
J Neurosci. 2005 Jun 1;25(22):5305-13. Erratum in: J Neurosci. 2005 Jun 29;25(26):6261-2.
14.Phosphorylated Extracellular Signal-regulated Kinases are Significantly Increased in Malignant Mesothelioma.
de Melo M, Gerbase MW, Curran J, Pache JC.
J Histochem Cytochem. 2006 Mar 3;
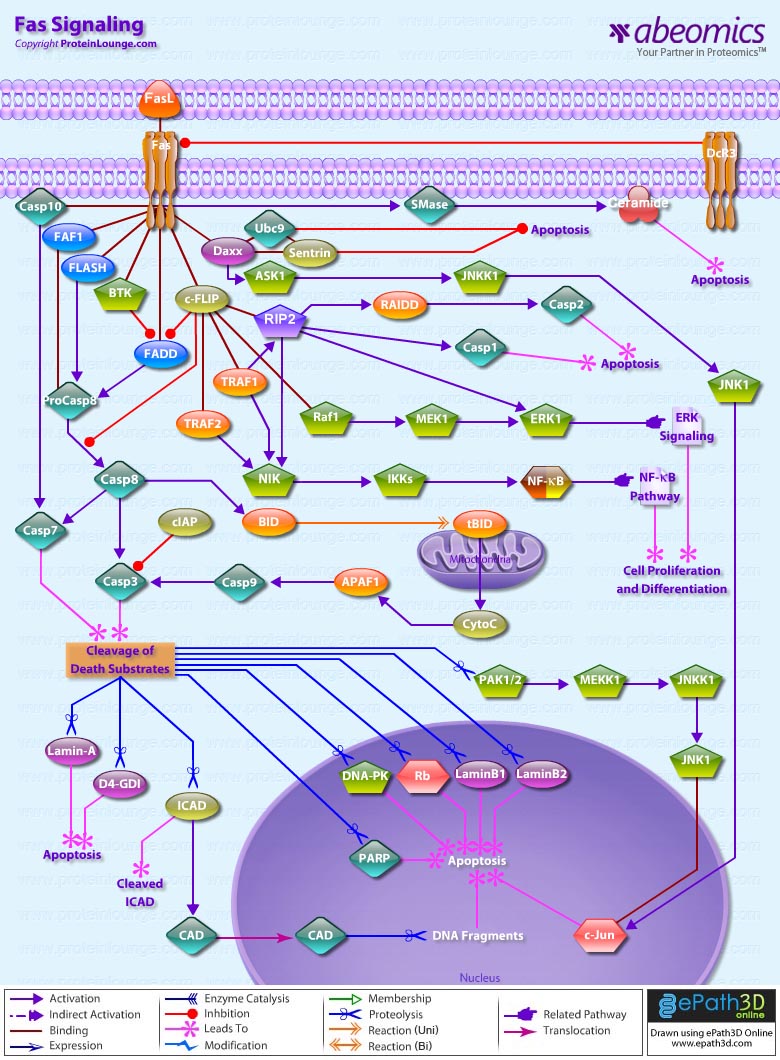
Fas (also called Apo1 or CD95) is a death domain-containing member of the TNFR (Tumor Necrosis Factor Receptor) superfamily. It has a central role in the physiological regulation of Programmed Cell Death and has been implicated in the pathogenesis of various malignancies and diseases of the immune system. Although the FasL (Fas Ligand)-Fas system has been appreciated mainly with respect to its death-inducing function, it also transduces proliferative and activating signals through pathways that are still poorly defined. The Fas Receptor induces an apoptotic signal by binding to FasL expressed on the surface of other cells. Fas is a Type-I transmembrane protein, where as FasL a Type-II Transmembrane protein of TNF family and can be shed in a soluble form by action of metalloproteinase (Ref.1).
The Fas Receptor upon binding to the FasL trimerizes and induces Apoptosis through a cytoplasmic domain called DD (Death Domain) that interacts with signaling adaptors like FAF-1 (Fas-Associated Factor-1), FADD (Fas-Associated Death Domain), Daxx, FAP-1, FLASH (FLICE-associated huge) and RIP (Receptor-Interacting Protein). FADD carries a DED (Death Effector Domain) and by homologous interaction it recruits the DED containing Procaspase8 protein which is in inactive state. This protein complex is also known as DISC (Death-Inducing Signaling Pathways) and exists in Type-I cells. Procaspase8 is proteolytically activated to Caspase8. FADD also helps in the activation of Caspase10. Upon activation, Caspase8 and Caspase10 cleave and activate downstream effector Caspases, including Caspase3, 6 and 7. Activated Caspase8 activates Caspase3 through two pathways; The complex one is that Caspase8 cleaves BID (Bcl2 Interacting Protein) and its COOH-terminal part translocates to mitochondria where it triggers the release of mitochondrial pro-apoptotic factors like CytoC (Cytochrome-C) and SMAC (Second Mitochondria-derived Activator of Caspases) also called Diablo. The released CytoC binds to APAF1 (Apoplectic Protease Activating Factor-1) together with dATP and Procaspase9 and activates Caspase9. Caspase9 cleaves Procaspase3 and activates Caspase3. Another pathway is that Caspase8 cleaves Procaspase3 directly and activates it. Both pathways are regulated at the level of Caspase8 activation by the endogenous inhibitor FLIP (FLICE (FADD Like IL-1Beta-Converting Enzyme)-Inhibitory Protein), which may also be recruited by FADD. Interestingly, FLIP may also participate in an alternate signalling pathway, recruiting TRAF1 (Tumor Necrosis Factor-Associated Factor-1), TRAF2 (Tumor Necrosis Factor-Associated Factor-1), the MAPKKK (MAP Kinase Kinase Kinase) Raf1 and RIP to activate ERK (Extracellular signal-Regulated Kinase) and NF-KappaB (Nuclear Factor-KappaB) pathways, leading to proliferation and/or inflammation. This differential activity of FLIP, which appears to reflect activity of short vs long FLIP isoforms that promote the death vs proliferation pathways, respectively, mediates a major decision in response to Fas signalling: death vs proliferation/inflammation. Caspases are inhibited by IAPs (Inhibitor of Apoptosis Proteins) (Ref.2 & 3).Caspase3, 6 and 7 once activated lead to breakdown of several cytoskeletal and nuclear proteins (structural, signaling proteins or kinases) like GDID4 (GDP-Dissociation Inhibitor-D4), PARP (Poly ADP-Ribose Polymerase), Alpha-Fodrin, GAS2 (Growth Arrest Specific-2) Lamin-A and B (B1 and B2) and PAK (p21-Activated Kinase) thus inducing apoptosis. PAK activity resulting from Caspase-mediated cleavage is a necessary component of JNK (c-Jun Terminal Kinase) activation induced by Fas receptor signaling and thus PAK can also contribute to the induction of cell death. Caspase3 also cleaves ICAD, the inhibitor of CAD (Caspase Activated DNAse), which frees CAD to enter the nucleus and cleave DNA. In the nucleus, activated Caspase3 also cleaves DNA-PKcs and release the cleaved fragments in the cytosolic compartment. Stimulation of Fas also results in significant alterations of Rb (Retinoblastoma protein) (Ref.4 & 5).
Besides the FADD/Caspase8 signaling cascade, a number of other signaling pathways are also activated by the Fas Receptor. Among the molecules involved in Fas-mediated signaling, Daxx and RIP have been shown to bind to the FasR, modulating its signal. Daxx (Death-Domain Associated protein) binds to the Fas death domain although Daxx itself does not contain a DD. Daxx and FADD bind independently to Fas and activate distinct pathways. Daxx can enhance Fas-mediated apoptosis by activating the JNK kinase cascade, culminating in the phosphorylation and activation of transcription factors such as c-Jun. The JNK kinase kinase ASK1 (Apoptosis Signal Regulating Kinase 1) sequesters Daxx in the cytoplasm. After Fas stimulation, Daxx interacts with and activates ASK1. In the absence of ASK1, Daxx is found in the nucleus, where it localizes to PODs (PML (Promyelocytic Leukemia Protein) Oncogenic Domains). ASK1 and Daxx can induce a Caspase-independent form of cell death that is independent from the kinase activity of ASK1. ASK1 is also able to induce Caspase-dependent apoptosis under critical requirement of its kinase activity. Daxx also interact with Sentrin, a ubiquitin-like protein that can covalently modify cellular proteins, and is Fas binding protein that protects cells against Fas induced cell death. Also, Daxx interacted with Ubc9, an essential protein as a key-conjugating enzyme. Colocalization of Fas, Sentrin, and Ubc9 binding regions suggests the importance of that region upon the regulation of Daxx. In addition, FLASH enhances the activation of Caspase8 in Fas-mediated apoptosis. Thus, DED-containing proteins seem to modulate the apoptotic process. FAF1 is a Fas-associating molecule, which enhances Fas mediated apoptosis. The N-terminus of FAF1 binds to the DD of Fas even though it does not contain the typical death domain. FAF1 is a component of Fas-DISC, and DISC is formed by interaction of the DED-like region (amino acid 181–381 of FAF1) of FAF1 and the DEDs of Caspase8 and FADD (Ref.6, 7, 8 & 9).
The Receptor Interacting Proteins (RIP, RIP2, RIP3, RIP4) are death domain-containing proteins which, uniquely, possess serine/threonine kinase activity. RIP associates with the Fas death domain but can also interact directly with FADD and FLIP. RIP overexpression results in NF-KappaB translocation, JNK activation, and apoptosis. RIP family proteins may transduce non-apoptotic signal through several pathways, including NF-KappaB and ERK. NF-KappaB activation by RIP is dependent on functional NIK (NF-KappaB-Inducing Kinase), and may involve NIK recruitment into the DISC. RIP mediated activation of NF-KappaB, while RIP is bound to FLIP, may account at least in part for the observation that FLIP can regulate NF-KappaB activation. Activation of NF-KappaB results in its translocation to the nucleus, where it acts as a transcription factor. NF-KappaB activation usually induces proliferation, differentiation, or inflammation, but may also promote apoptosis, perhaps accounting for the discrepancies in reported RIP functions in different cellular contexts. Activated RIP2 can phosphorylate ERKs (Extracellular signal-Regulated Kinases), thus activating the ERK pathway independently of MEK. Finally, RIP can also activate Caspase1 directly, and Caspase2 via the adaptor protein RAIDD (RIP-associated ICH-1/Ced3-homologous protein with a Death Domain). Activation of Caspase1 allows the processing of IL-1Beta (pro-Interleukin-1Beta) to active IL-1Beta, a potent inducer of inflammation. Thus, RIP can mediate Fas-induced differentiation and inflammation through a number of different pathways. An acidic SMase (Sphingomyelinase) is also activated in response to stimulation of Fas, resulting in the production of Ceramide, a mediator of cell stress and well known stimuli of apoptosis (Ref.2 & 10).
In B-Cells, BTK (Bruton’s Tyrosine Kinase) associates with the death receptor Fas and impairs its interaction with FADD, which is essential for the recruitment and activation of FLICE by Fas during the apoptotic signal, thereby preventing the assembly of DISC after Fas-ligation. Fas pathway can also be regulated at the transcriptional level, such as by the tumour suppressor p53. p53 positively regulates genes, including Fas and Bax that are involved in FasL-induced apoptosis. FasL has been implicated in maintaining immune privileged sites such as the eye, testis, brain, joints, and pregnant uterus by inducing apoptosis in activated infiltrating leukocytes and lymphocytes that express Fas. Since, Fas and FasL are known regulators of apoptosis in cells of the immune system, blocking Fas/FasL interactions in human colon carcinoma cells can lead to thymine less death. Fas/FasL interaction may also be involved in target destruction during organ-specific autoimmune diseases, such as Hashimoto’s Thyroiditis, Insulin-dependent Diabetes Mellitus and Multiple Sclerosis (Ref.11 & 12).
Malhi H, Guicciardi ME, Gores GJ.
Physiol Rev. 2010 Jul;90(3):1165-94.2.Cellular stress responses: cell survival and cell death.
Fulda S, Gorman AM, Hori O, Samali A.
Int J Cell Biol. 2010;2010:214074. Epub 2010 Feb 21.3.Receptor-interacting protein (RIP) kinase family.
Zhang D, Lin J, Han J.
Cell Mol Immunol. 2010 Jul;7(4):243-9. Epub 2010 Apr 12.
4.Apoptosis: a review of programmed cell death.
Elmore S.
Toxicol Pathol. 2007 Jun;35(4):495-516.
5.Caspases and kinases in a death grip.
Kurokawa M, Kornbluth S.
Cell. 2009 Sep 4;138(5):838-54.
6.Targeting Fas in osteoresorptive disorders.
Kovacic N, Grcevic D, Katavic V, Lukic IK, Marusic A.
Expert Opin Ther Targets. 2010 Oct;14(10):1121-34.
7.The roles of ASK family proteins in stress responses and diseases.
Hattori K, Naguro I, Runchel C, Ichijo H.
Cell Commun Signal. 2009 Apr 24;7:9.
8.Daxx is a transcriptional repressor of CCAAT/enhancer-binding protein beta.
Wethkamp N, Klempnauer KH.
J Biol Chem. 2009 Oct 16;284(42):28783-94. Epub 2009 Aug 18.
9.PIAS1 interacts with FLASH and enhances its co-activation of c-Myb.
Alm-Kristiansen AH, Lorenzo PI, Molværsmyr AK, Matre V, Ledsaak M, Sæther T, Gabrielsen OS.
Mol Cancer. 2011 Feb 21;10:21.
10.Integration of Apoptosis and Metabolism.
Yi CH, Vakifahmetoglu-Norberg H, Yuan J.
Cold Spring Harb Symp Quant Biol. 2011 Nov 16. [Epub ahead of print]
11.a-TEA cooperates with chemotherapeutic agents to induce apoptosis of p53 mutant, triple-negative human breast cancer cells via activating p73.
Tiwary R, Yu W, Sanders BG, Kline K.
Breast Cancer Res. 2011 Jan 7;13(1):R1.
12.The tumour necrosis factor/TNF receptor superfamily: therapeutic targets in autoimmune diseases.
Vinay DS, Kwon BS.
Clin Exp Immunol. 2011 May;164(2):145-57. doi: 10.1111/j.1365-2249.2011.04375.x. Epub 2011 Mar 14.

ESCs (Embryonic Stem Cells) are Pluripotent cells capable of differentiating into any cell type of the body. Only three species of Mammals have yielded long-term cultures of self-renewing ESCs- Mice, Monkeys, and Humans. Human ESCs are derived from Blastocysts, multicellular structures originating from four cleavages of fertilized oocytes. Isolated from the ICM (Inner Cell Mass) of Blastocysts, the ESCs retain properties of self-renewal and the potential to be committed and to differentiate toward most cell lineages. They are able to spontaneously give rise to different progenies of the three embryonic layers, namely, the Ectoderm, the Mesoderm and the Endoderm. The Pluripotency of ESCs has attracted great attention for their potential use in tissue and cell therapy. However, the molecular and developmental mechanisms controlling Pluripotency and Differentiation of ESCs are largely unknown. Human ESC Pluripotency is regulated by a combination of Extrinsic and Intrinsic factors. Unlike Mouse, Extrinsic factor LIF (Leukemia Inhibitory Factor) is not sufficient to maintain Human ESC and BMPs (Bone Morphogenic Proteins) cause rapid differentiation. Instead, FGF (Fibroblast Growth Factor) signaling and a balance between TGF-Beta (Transforming Growth Factor-Beta) /Activin and BMP signaling are central to the self-renewal of Human ESCs. Intrinsic factors regulating Pluripotency in Human ESCs include a battery of transcription factors including Oct4 (Octamer Binding Transcription Factor-4), SOX2 (SRY (Sex Determining Region-Y) Box-2) and Nanog (Ref.1 & 2).
TGF-Beta superfamily of ligands, which contains about 40 potential ligands in the Human genome, play a major role maintaining the Self-renewing Human ESCs. They signals through two main branches: the SMAD1/5 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-1/5) branch, which transduces on behalf of BMP and GDF (Growth Differentiation Factor) ligands via the Type I receptors ALK1 (Activin Receptor-Like Kinase-1), ALK2 (Activin Receptor-Like Kinase-2), ALK3 (Activin Receptor-Like Kinase-3) and ALK6 (Activin Receptor-Like Kinase-6); and the TGF-Beta/Activin/Nodal branch, which involves the activation of SMAD2/3 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-2/3) via ALK4 (Activin Receptor-Like Kinase-4), and ALK5 (Activin Receptor-Like Kinase-5) and ALK7 (Activin Receptor-Like Kinase-7). There are also two inhibitory SMADs – SMAD6 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-6), which selectively inhibits SMAD1/5; and SMAD7 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-7), which inhibits both branches of TGF-Beta signaling, that provide a repressive input on the pathway. Upon activation by phosphorylation and association with a common SMAD4 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-4), the receptor-activated SMADs translocate to the nucleus and, in concert with other transcription factors, regulate gene expression. Activation of the TGF-Beta/Activin/Nodal branch through SMAD2/3 is associated with Pluripotency and is required for the maintenance of the undifferentiated state in Human ESCs. SMAD2/3 pathway is also required for positive regulation of several factors of TGF-Beta signaling. These factors include Nodal, Cripto, LEFTY1 (Left-right determination factor-1) and LEFTY2 (Left-right determination factor-2). Cripto encodes an EGF-CFC co-receptor that is essential for responsivity to Nodal, and LEFTY1 and LEFTY2 are both inhibitors of Nodal signaling. Expression of Nodal, LEFTY1 and LEFTY2 remain high in undifferentiated Human ESCs and reduce upon differentiation (Ref.3, 4 & 5).
In contrast to TGF-Beta/Activin/Nodal signaling, which promotes the maintenance of Pluripotent Human ESCs, BMP signaling is unable to support self-renewal and is associated with differentiation to Trophoblast or Extraembryonic Endoderm cells. In Human ESCs, BMP4 (Bone Morphogenic Protein-4) induces differentiation into Mesoderm and Ectoderm, whereas BMP2 (Bone Morphogenic Protein-2) promotes Extraembryonic Endoderm differentiation. Repression of BMP signaling in Human ESCs by Noggin and FGF supports long term Self-renewal. FGF2 (Fibroblast Growth Factor-2) is known as the best known factor promoting Self-renewal in Human ESCs. Exogenous FGF2 is capable of maintaining Human ESCs in the absence of serum and feeder cells. Binding of FGF to its receptor and Heparin leads to receptor autophosphorylation and activation of intracellular signaling cascades, including the Ras/ERK (Extracellular Signal-Regulated Kinase) pathway, the PLC-Gamma (Phospholipase-C-Gamma)/Ca2+ (Calcium) pathway, and the PI3K (Phosphoinositide 3-Kinase) pathway. FGF2 promote self-renewal of Human ESCs by activating the PI3K pathway. PI3K catalyzes the conversion of PIP2 (Phosphatidylinositol (3,4)-bisphosphate) to PIP3 (Phosphatidylinositol (3,4,5)-trisphosphate). The binding of PIP3 to the PH domain anchors Akt to the Plasma membrane and allows its phosphorylation and activation by PDK-1 (Phosphoinositide-Dependent Kinase-1). Activated Akt promotes cell proliferation, survival, growth, and motility and is implicated in tumorogenicity. Survival of cell leads to Pluripotency. FGF2- dependent PI3K/Akt activation is required for the efficient expression of ECM (Extracellular Matrix) proteins. Role of PI3K in maintaining Human ESC self-renewal requires further investigation. A role for PI3K has been proposed in Human ESC survival, where it mediates the survival activity of Neurotrophins, signaling through TRK Receptors (Tyrosine Kinase Receptors) (Ref.3, 6, 7 & 8).
WNTs (Wingless-Type MMTV Integration Site Family Members) proteins are also believed to play an important role in controlling ESC maintenance. Canonical WNT signaling involves the binding of WNT ligands to the Frizzled receptors. This, in turn, activates Dsh (Dishevelled), which displaces GSK-3Beta (Glycogen Synthase Kinase-3Beta) from the APC (Adenomatous Polyposis Coli) /AXIN (Axis Inhibitor) complex, preventing Ubiquitin-mediated degradation of Ctnn-Beta (Catenin-Beta). Subsequently, Ctnn-Beta accumulates and translocates into the nucleus where it associates with TCF (T Cell Factor)/ LEF (Lymphoid Enhancer Factor) proteins to activate transcription of WNT target genes. WNT signaling is known to be involved in regulating the proliferation of Stem cells, including Intestinal and Hematopoietic cells, and the Self-renewal of Hematopoietic stem cells. Components of the WNT signaling pathway are present in Human ESCs, although levels of different receptors varied between undifferentiated and differentiated populations. WNT is believed to stimulate Human ESC proliferation as well as differentiation (Ref.3, 9 & 10).
S1P (Sphingosine-1-Phosphate), a bioactive Lysophospholipid, also supports Human ESC self-renewal. S1P signals both extracellularly, through EDG (Endothelial Differentiation Gene) receptors coupled to G-Proteins, and intracellularly by undefined mechanisms. S1P has been implicated in a diverse range of biological processes, including cell growth, differentiation, migration and apoptosis in many different cell types. Because the prevention of apoptosis is a common self-renewal mechanism, S1P potentially aids the self-renewal process in Human ESCs cells. PDGF (Platelet Derived-Growth Factor) has also been implicated in the prevention of apoptosis. PDGF promotes intracellular S1P signaling by activating SphK (Sphingosine Kinase), which in turn converts Sphingosine to S1P. The combination of Extracellular PDGF and S1P supports Human ESC self-renewal (Ref.11 & 12).
The undifferentiated state of Human ESCs is also maintained by the action of transcription factors some of which are ESC-specific and common between Human and Mouse ESCs. Major transcription factors regulating Pluripotency include Oct4, SOX2 and Nanog. The best-characterized gene of these is Oct4, which functions to maintain Pluripotency both in vivo and in vitro. Oct4 is a POU domain transcription factor that is specifically expressed in all Pluripotent cells. There is scarce knowledge concerning the upstream factors that regulate the expression of the key transcription factors. The expression of Oct3/4 is regulated by a proximal enhancer and promoter in the Epiblast and by a distal enhancer and promoter at all other stages in the Pluripotent cell lineage. Limited downstream target genes of Oct3/4 have been identified, out of which FGF4 is the most common target. SOX2, a Sry-related transcription factor, activates the transcription of target genes such as FGF4, in cooperation with Oct3/4. SOX2 expression is controlled by Oct4 and SOX2, suggesting a positive feedback mechanism that is related to the maintenance of ESC self-renewal. Two regulatory elements, SRR1 and SRR2, have been found in the area of the SOX2 gene in undifferentiated ES cells. In the presence of these two regulatory elements, a high level of gene expression is achieved through synergistic activity. The partnership between Oct3/4 and SOX2 is involved in the expression of the UTF1 (Undifferentiated embryonic cell Transcription Factor-1) gene found to encode an ESC-specific co-activator. The binding sites for both factors are located in the second intron of the UTF1 gene with no intervening spaces. Thus, Oct3/4 SOX2 partnership is indispensable in the maintenance of Pluripotency. Stellar, a gene with similar expression to Oct3/4 has recently been identified. Its expression is mainly restricted to Pluripotent Human ESCs, Premeiotic Germ cell tumors, and Testicular Germ cell tumors (Ref.13, 14 & 15).
Nanog is another member of the group of transcription factors whose functions are deemed essential for the process of Self-renewal in Human ESCs. Nanog is a NK2-family homeobox transcription factor and it acts by transcriptional activation, achieved by binding to homeobox domains in downstream target genes. Analogous to Oct3/4 and SOX2, Nanog expression is high in Human ESCs and is down regulated as cells differentiate. Transcription of Nanog is regulated by the binding of Oct3/4 and SOX2 to the Nanog promoter. Oct3/4, SOX2, and Nanog co-occupy the promoters of a large range of genes, many of which encode developmentally important transcription factors. Oct3/4, SOX2, and Nanog together occupy a minimum of 353 of Human ESC genes to maintain Pluripotency. Thus, Human ESCs exhibit a number of signaling pathways involved in Self-renewal and Pluripotency that are interdependent and display a range of cross-talk mechanisms. Deciphering these pathways holds promise for generating improved methodologies for maintenance and proliferation of Pluripotent Human ESCs in vitro. Together, the understanding of the exogenous and endogenous factors determining cell fate will facilitate the use of these cells in cell-based therapies and will allow understanding of early developmental processes (Ref.2, 16, 17 & 18).
Biswas A, Hutchins R.
Stem Cells Dev. 2007 Apr;16(2):213-22.2.Human embryonic stem cells: the battle between self-renewal and differentiation.
Darr H, Benvenisty N.
Regen Med. 2006 May;1(3):317-25.3.Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells.
Xiao L, Yuan X, Sharkis SJ.
Stem Cells. 2006 Jun;24(6):1476-86.
4.Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3.
Besser D.
J Biol Chem. 2004 Oct 22;279(43):45076-84.
5.TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells.
James D, Levine AJ, Besser D, Hemmati-Brivanlou A.
Development. 2005 Mar;132(6):1273-82.
6.BMPs regulate differentiation of a putative visceral endoderm layer within human embryonic stem-cell-derived embryoid bodies.
Conley BJ, Ellis S, Gulluyan L, Mollard R.
Biochem Cell Biol. 2007 Feb;85(1):121-32.
7.Noggin and bFGF cooperate to maintain the pluripotency of human embryonic stem cells in the absence of feeder layers.
Wang G, Zhang H, Zhao Y, Li J, Cai J, Wang P, Meng S, Feng J, Miao C, Ding M, Li D, Deng H.
Biochem Biophys Res Commun. 2005 May 13;330(3):934-42.
8.Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells.
Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA.
Nat Methods. 2005 Mar;2(3):185-90.
9.Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency.
Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, Kahn M.
Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5668-73.
10.Wnt3a regulates survival, expansion, and maintenance of neural progenitors derived from human embryonic stem cells.
Davidson KC, Jamshidi P, Daly R, Hearn MT, Pera MF, Dottori M.
Mol Cell Neurosci. 2007 Aug 7;
11.Development of serum-free culture systems for human embryonic stem cells.
Chase LG, Firpo MT.
Curr Opin Chem Biol. 2007 Aug;11(4):367-72.
12.Essential roles of sphingosine-1-phosphate and platelet-derived growth factor in the maintenance of human embryonic stem cells.
Pébay A1, Wong RC, Pitson SM, Wolvetang EJ, Peh GS, Filipczyk A, Koh KL, Tellis I, Nguyen LT, Pera MF.
Stem Cells. 2005 Nov-Dec;23(10):1541-8. Epub 2005 Aug 4.
13.Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells.
Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J.
Stem Cells. 2007 Feb;25(2):500-10.
14.A protein interaction network for pluripotency of embryonic stem cells.
Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH.
Nature. 2006 Nov 16;444(7117):364-8. Epub 2006 Nov 8.
15.A UTF1-based selection system for stable homogeneously pluripotent human embryonic stem cell cultures.
Tan SM, Wang ST, Hentze H, Droge P.
Nucleic Acids Res. 2007 Sep 12;
16.Nanog and transcriptional networks in embryonic stem cell pluripotency.
Pan G, Thomson JA.
Cell Res. 2007 Jan;17(1):42-9.
17.NANOG maintains self-renewal of primate ES cells in the absence of a feeder layer.
Yasuda SY, Tsuneyoshi N, Sumi T, Hasegawa K, Tada T, Nakatsuji N, Suemori H.
Genes Cells. 2006 Sep;11(9):1115-23.
18.Core transcriptional regulatory circuitry in human embryonic stem cells.
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA.
Cell. 2005 Sep 23;122(6):947-56.
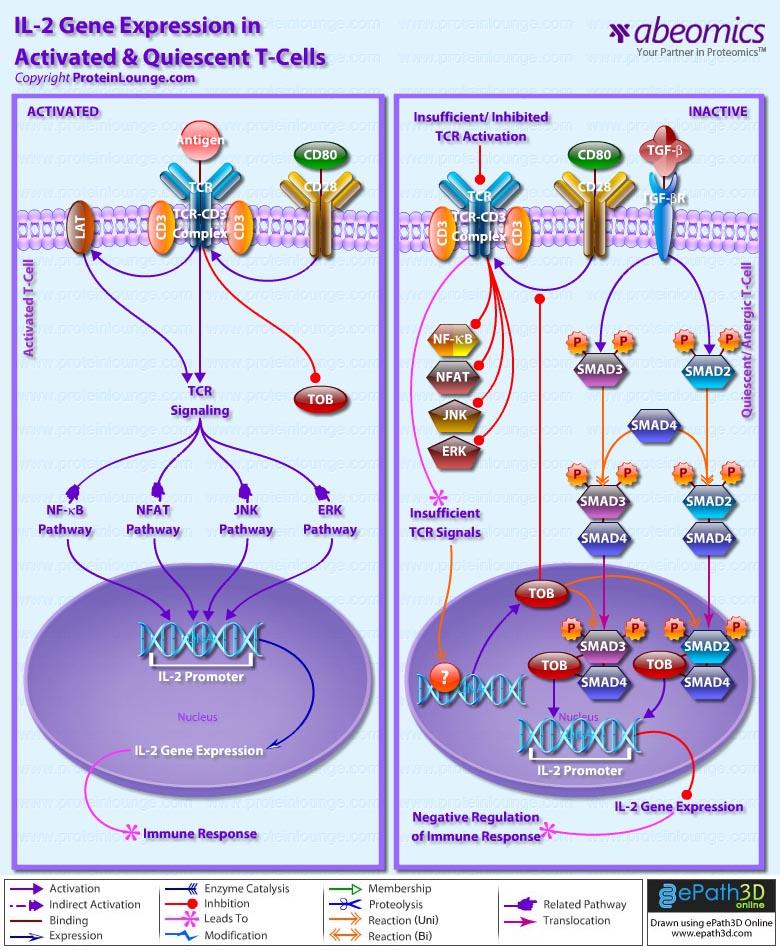
IL-2 (Interleukin-2) is a biological response modifier (cytokine), which stimulates the growth, proliferation and subsequent differentiation of disease-fighting blood cells, like T-Cells, B-Cells, NK (Natural Killer) cells, monocytes, macrophages, and oligodendrocytes. It is a powerful immunoregulatory lymphokine that was originally described as “T-Cell growth factor” and is secreted primarily by Antigen-activated T-Cells (Ref.1). Human IL-2 is a 133-amino acid polypeptide with a molecular mass of 15-18 kDa. In an autocrine fashion, the Antigen-primed THC (T Helper Cell) secretes IL-2, stimulating itself as well as other neighboring Antigen-primed T-Cells to proliferate (T-Cell activation and proliferation). Growing T-cells in long term culture require IL-2 as a growth factor, and this cytokine is crucial for achieving T-cell-mediated immunity, and thus, regulation of the immune response (Ref.2).
In normal T-Cells, engagement of TCR (T-Cell Receptor)-CD3 complexes, costimulation by CD28 and recruitment of LAT (Linker of Activated T-Cells) leads to a membrane-proximal cascade of tyrosine phosphorylation events that ultimately lead to the activation of multiple pathways, including ERK (Extracellular Signal Regulated Kinase), JNK (c-Jun N-terminal Kinase), NF-KappaB (Nuclear Factor-KappaB) and NFAT (Nuclear Factor of Activated T-cells) leading to the activation of the IL-2 gene promoter region subsequently followed by IL-2 gene expression (Ref.3). Since IL-2 gene expression in T-Cells plays various critical roles in orchestrating the immune responses, deregulation of T-Cell function, whether by defect or by excess, results in dire consequences for the organism i.e., immunodeficiency and autoimmunity (Ref.1). If the T-Cell response is too great, it will culminate in excessive expression of IL-2, resulting in hyperactivation of immune response, that may give rise to autoimmune disorders or tissue injury. Therefore, regulation of T-Cell activation and maintenance of the T-Cells in quiescent and unresponsive state is an essential component of the balanced functioning of the immune system. Suboptimal cross-linking of the TCR by Antigen, which often happens under physiological conditions, is not sufficient to induce IL-2 gene expression, and thus, fails to induce a productive immune response, but instead leads to T-Cell quiescence (Ref.4).
The quiescent state of unstimulated T-lymphocytes may be due to a lack of activation signals or because of the presence of inhibition signals. In contrast to primary unstimulated T-Cells, which can enter the cell cycle and clonally expand after Antigen-specific stimulation, anergic T-Cells do not proliferate. Instead, they remain in a state of long-term Antigen-specific unresponsiveness, termed as T-Cell anergy. The quiescent state and the anergic state both result because of defective IL-2 gene expression. In quiescent T-Cells, the activation signals (Antigens) are either absent or, are insufficient to trigger a productive immune response. Presence of inhibition signals also gives rise to quiescence and anergy (Ref.2 & 4). The activation signals are not sufficient to trigger activation of the multiple pathways, including ERK, JNK, NF-KappaB and NFAT that usually lead to the activation of the IL-2 gene promoter region. Therefore, IL-2 gene transcription is hindered. On the other hand, the ERK, JNK, NF-KappaB, and NFAT pathways are inhibited by the presence of inhibitory substances, both in anergic, and quiescent states. TOB (Transducer of ERBB2), a negative regulator of IL-2 transcription and T-Cell proliferation—is constitutively expressed in unstimulated peripheral blood T-lymphocytes and selectively expressed in anergic T-Cells. TOB is a member of the TOB and BTG (B-Cell Translocation Gene) anti-proliferative protein family (Ref.5). Its expression is highest in unstimulated and anergic T-Cells, and is reduced in activated T-Cells. Once expressed, TOB inhibits the process of costimulation of TCR signaling by CD28, and thus, repression of pathways, that normally lead to the IL-2 expression. In activated T-Cells, expression of TOB is not normally observed. Otherwise, down-regulation of TOB is necessary for T-Cell activation. Repression of IL-2 gene by TOB is brought about by the interaction of TOB with SMAD proteins. TOB inhibits TCR-CD3 and CD28–mediated IL-2 gene transcription by augmenting the inhibitory effect of the SMAD proteins. TOB enhances SMAD binding to the -105 negative regulatory element of the IL-2 promoter. In T-Cells, SMADs mediate signals induced by TGF-Beta (Transforming Growth Factor-Beta) through its receptors: TGF-BetaRII and TGF-BetaRI. The activated TGF-BetaRs phosphorylate the downstream targets: SMAD2 and SMAD3, which form hetero-oligomeric complexes with SMAD4 and translocate to the nucleus. The SMAD complex downregulates IL-2 gene expression by binding to the -105 negative regulatory element of the IL-2 Promoter. TOB enhances the binding of the SMADs to the -105 negative regulatory element of the IL-2 promoter that represses IL-2 expression (Ref.6). Since IL-2 plays various critical roles in orchestrating the immune responses, a complete knowledge about its expression and repression would lead to an understanding of how the immune system could be better manipulated to overcome afflictions such as Cancer, Infection and Autoimmune diseases. Not only is it produced by mature T lymphocytes on stimulation but also constitutively by certain T-Cell lymphoma cell lines. IL-2 is the major tool in cytokine immunotherapy, where, prevention or reduction of disease progression is achieved through stimulation of cell-mediated immunity (i.e., immune reconstitution) by administration of an exogenous T helper cell cytokines. IL-2 has been used successfully to manage several human Cancers (Metastatic Melanoma, Acute Myelogenous Leukemia, and Metastatic Renal Cell Carcinoma); and has recently been shown to be effective at improving immune responses in patients with HIV1 (Human Immunodeficiency Virus Type-1) disease (Ref.7).
Osinalde N, Moss H, Arrizabalaga O, Omaetxebarria MJ, Blagoev B, Zubiaga AM, Fullaondo A, Arizmendi JM, Kratchmarova I.
J Proteomics. 2011 Dec 10;75(1):177-91. Epub 2011 Jun 23.2.Helper T cell IL-2 production is limited by negative feedback and STAT-dependent cytokine signals.
Villarino AV, Tato CM, Stumhofer JS, Yao Z, Cui YK, Hennighausen L, O’Shea JJ, Hunter CA.
J Exp Med. 2007 Jan 22;204(1):65-71. Epub 2007 Jan 16.3.Viral modulation of T-cell receptor signaling.
Jerome KR.
J Virol. 2008 May;82(9):4194-204. Epub 2008 Feb 20. Review. No abstract available.
4.Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells.
Sprent J, Surh CD.
Nat Immunol. 2011 Jun;12(6):478-84. Review.
5.Enhanced transgene expression in quiescent and activated human CD8+ T cells.
Cooper LJ, Topp MS, Pinzon C, Plavec I, Jensen MC, Riddell SR, Greenberg PD.
Hum Gene Ther. 2004 Jul;15(7):648-58.
6.Tob, a member of the APRO family, regulates immunological quiescence and tumor suppression.
Tzachanis D, Boussiotis VA.
Cell Cycle. 2009 Apr 1;8(7):1019-25. Epub 2009 Apr 30. Review.
7.Interleukin-2 immunotherapy and AIDS-related cytomegalovirus retinitis.
Dix RD, Cousins SW.
Curr HIV Res. 2004 Oct;2(4):333-42.

IL-3 (Interleukin-3) is a cytokine that regulates haemopoiesis, the formation of blood cells in the body. IL-3, also called multi-CSF (multi-lineage colony stimulating factor), is produced by T cells and mast cells, after activation with mitogens or antigens. The molecular weight of IL-3, a protein of 140 amino acids, ranges from 14 to 36 kDa. It stimulates eosinophils and B cell differentiation while it inhibits LAK (Lymphokine-Activated Killer) cell activity. IL-3 shares several biological activities with GM-CSF. IL-3 is capable of inducing the growth and differentiation of multi-potential haematopoetic stem cells, neutrophils, eosinophils, megakaryocytes, macrophages, lymphoid and erythroid cells (Ref.1). IL-3R (IL-3 receptor) is composed of two polypeptide chains, an Alpha subunit of 60-70kDa and a Beta subunit of 130-140kDa. Both subunits contain the extracellular conserved motifs found in the cytokine receptor superfamily. The common Beta -chain receptor subunit (c), which is shared among the receptors for IL-3, IL-5 and granulocyte/macrophage colony-stimulating factor, is essential for signal transduction in cell proliferation and differentiation induced by these cytokines, and plays a major role in recruiting intracellular signaling molecules such as JAK2 (Janus Kinase2) tyrosine kinase and STAT5 (Signal Transducers And Activators Of Transcription) (Ref.2).
The Alpha and Betac chains of the IL-3 receptor are not associated in the absence of ligand. The presence of ligand induces alpha and Betac chain association to form a heterodimeric complex. Receptor activation is followed by activation of receptor associated JAK2 kinase and tyrosine and serine phosphorylation of the Betac chain cytoplasmic tail. Activation of JAK2 leads to phosphorylation of the IL-3R Betac chain on multiple tyrosine residues which in turn serve as docking sites for other signal transducing proteins, the most important of which are STATs. IL-3 activation of hematopoetic cells appears to lead to the activation of multiple STATs, which includes STAT1, STAT3, STAT5 and STAT6. In addition, stimulation with IL-3 leads to morphological changes of cells through tyrosine phosphorylation of Betac and its associated protein pp90. In unstimulated cells, ectopically expressed RON tyrosine kinase localizes with the IL-3 receptor Betac (Ref.3).
Src family kinases mediate the phosphorylation of STAT3 mediated by IL-3/receptor interactions and play a critical role in signal transduction pathways associated with myeloid cell proliferation. One or both isoforms of STAT5 interact directly with JAK2, which in turn mediates their phosphorylation, STAT3 activation require its interaction with c-Src, which in turn mediates its phosphorylation. In addition to the activation of STATs, IL-3 activates multiple signal transduction pathways, which includes the Ras and PI3K (Phosphatidylinositol-3 Kinase) pathways. Upon IL-3 stimulation, the adapter molecule SHC (Src Homology 2 Domain Containing) transforming protein is rapidly phosphorylated and associates with the phosphorylated Betac subunit of IL-3. IL-3 stimulation also results in tyrosine phosphorylation of the inositol phosphatase SHIP (SH2-Containing Inositol Phosphatase), which forms a complex with SHC, GRB2 (Growth Factor Receptor-Bound Protein 2) and SOS (Son Of Sevenless). This is followed by the activation of Ras and c-Raf, which results in downstream activation of ERK1 and ERK2 (Extracellular Signal-Regulated Kinases). Activation of the cascades culminates in the increased expression of transcription factors c-Jun and c-Fos. In addition to activation of ERKs, IL-3 also activates p38 and JNK. IL-3 induces a rapid activation of the lipid kinase PI3K. Downstream proteins recruited by the PI3K pathways upon IL-3 stimulation includes the Akt protein. Another downstream protein activated in response to IL-3 stimulation is p70S6k (p70 Ribosomal S6 Kinase), which also mediates its effect via interaction with Betac chain. Another protein that feeds into the PI3K-PKB/AKT pathway is the Cbl protein, which also docks onto the adaptor protein GRB2 and SHC. The BCL2 family mediates the cell survival function of IL-3. BCL2 and BCLXL are rapidly induced by IL-3, which depends upon JAK2 activation. IL-3 also regulates the glycolytic pathway. In Baf-3 cells IL-3 starvation leads to a decrease in glucose uptake and in lactate production. It is found that the eosinophils activated by IL-3 may contribute to T cell activation in allergic and parasitic diseases by presenting superantigens and peptides to T-Cells (Ref.4).
Celestin J, Rotschke O, Falk K, Ramesh N, Jabara H, Strominger J, Geha RS.
J Immunol. 2001 Dec 1;167(11):6097-104.2.Inhibition of GM-CSF/IL-3/IL-5 signaling by antisense oligodeoxynucleotides targeting the common beta chain of their receptors.
Allam M, Renzi PM.
Antisense Nucleic Acid Drug Dev. 2001 Oct;11(5):289-300.3.IL-3 signaling and the role of Src kinases, JAKs and STATs: a covert liaison unveiled.
Reddy EP, Korapati A, Chaturvedi P, Rane S.
Oncogene. 2000 May 15;19(21):2532-47.
4.Activation of the interleukin-3 gene by chromosome translocation in acute lymphocytic leukemia with eosinophilia.
Meeker, T. C.; Hardy, D.; Willman, C.; Hogan, T.; Abrams, J.
Blood 76: 285-289, 1990.
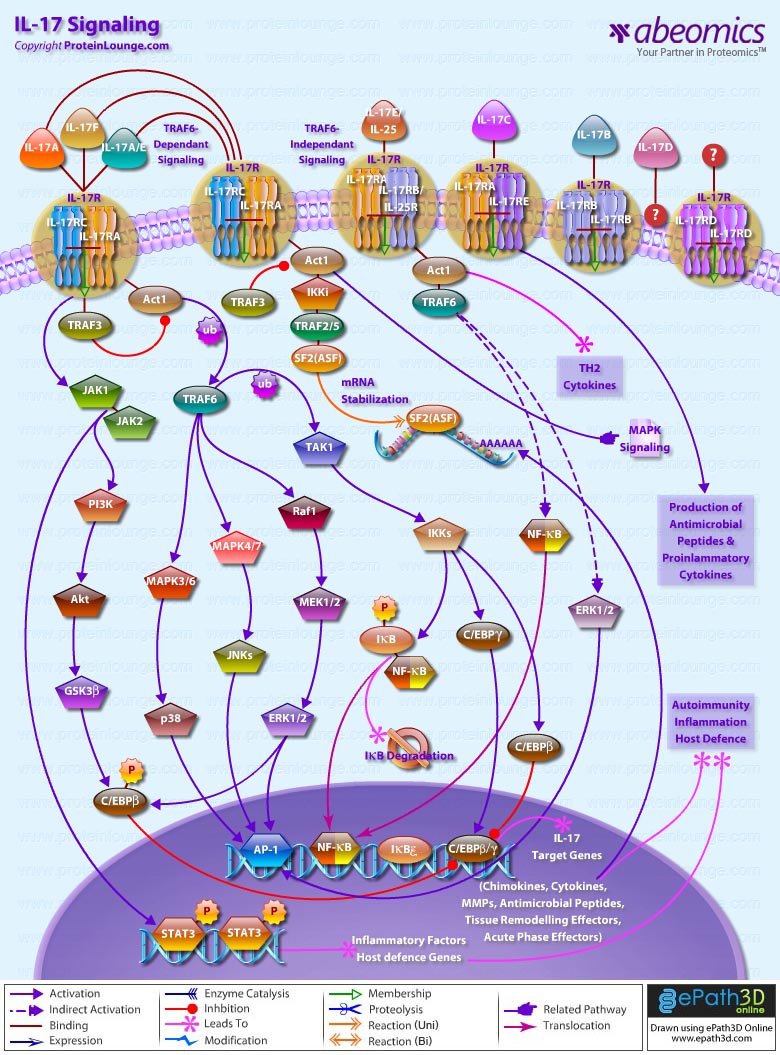
Cytokines are key messenger molecules in cell-to-cell communication and are involved in various aspects of the immune system such as maintaining homeostasis and mediating and resolving pathologic conditions. IL-17 (Interleukin-17) family is a group of cytokines sharing homology in amino acid sequences with highly conserved cysteine residues critical to their 3-dimensional shape. IL-17, the signature cytokine secreted by Th17 cells, is required for host defence against extracellular bacterial and fungal infections, and contributes to the pathogenesis of various autoimmune inflammatory diseases. IL-17 has become an important target for treating different forms of inflammatory disorders. Recent clinical trials with agents that target IL-17/IL-17R, the upstream regulation pathways of IL-17 expression and the downstream signaling pathways of IL-17 hold promise for treating autoimmune diseases. So far, six members, IL-17A (commonly refer to IL-17), IL-17B, IL-17C, IL-17D, IL-17E (also called IL-25) and IL-17F, have been identified. The IL-17R family contains five receptor subunits, IL-17RA–IL-17RE. Both IL-17 members and IL-17Rs have little homology to other known cytokines or cytokine receptors, and are classified as new cytokine and cytokine receptor families.IL-17 signals through a heteromeric receptor complex formed by IL-17R (IL-17RA) and IL-17RC, which are single-pass transmembrane proteins expressed by a variety of cells including astrocytes and microglia. IL-17RA can be bound by both IL-17A and IL-17F, but with 10-fold more affinity for IL-17A than for IL-17F. While both IL-17E and IL-17B bind IL-17RB, to induce Th2 cytokines, the ligand for IL-17RD is still unknown. IL-17C may be the ligand for IL-17RE, but the functional link needs to be identified. Binding of the first IL-17 receptor subunit to the ligand modulates the affinity and specificity of the second-binding event, thereby promoting heterodimeric versus homodimeric complex formation. Three remaining cytokines in the IL-17 family, IL-17B, IL-17C and IL-17D, currently are poorly studied with regards to their biological function and receptors.IL-17C which binds to the IL-17RE–IL-17RA heterodimeric complex is also important for host defence and autoimmunity (Ref.1, 2, 3&4).
IL-17A is a founding member of the IL-17 family and serves as an essential player in host defense during infection while aberrant expression of IL-17A is associated with many autoimmune diseases and cancer. IL-17A is a pleiotropic cytokine that acts on multiple cell types to enhance the production of proinflammatory molecules. IL-17 binds to the IL-17RA or IL-17RC as homodimers or heterodimers. Once IL-17F is secreted from CD4 and CD T-cells, it is recognized by IL-17RA/RC heteromeric complex. IL-17RA, then, recruits the adaptor molecule, Act1. Act1 activates TRAF6 as well as other transcription factors, leading to induction of cytokines, chemokines and MMPs (matrix metalloproteinase) genes. In addition, the presence of IL-17A/F heterodimers substantially influences the potency, specificity and the spectrum of the activity of these cytokines. Therefore, future studies should be directed to understand the mechanism and role of IL-17F and its related cytokines and provide a platform for development of therapeutic intervention for inflammatory diseases (Ref.5, 6, 7&8) .IL-17 has been shown to activate many common downstream signalling pathways, including NF-KappaB (Nuclear Factor-KappaB), the MAPKs: JNK (c-Jun N-terminal Kinase), p38 and ERK (Extracellular-signal regulated kinase), C/EBPbeta/gamma, PI3K and JAK/STATs. Signaling by PI3K through Akt leads to up-regulation of IL-17RA. Akt in turn phosphorylates a number of downstream targets that cumulatively serve to promote cell survival, including GSK3-Alpha/Beta (Ref.9&10). Another important function of IL-17 is that it can stabilize the mRNA of some proinflammatory cytokines and chemokines induced by TNF (Tumor Necrosis Factor). TRAF3 (TNF Receptor-Associated Factor-3) is a proximal negative regulator of the IL-17R and suppresses IL-17-mediated downstream events through interfering with the formation of the receptor signaling activation complex IL-17R–Act1–TRAF6 (Ref.2, 3, 11&12).
A unique TRAF6-independent signaling pathway activated by IL-1 .IKKi is recruited to the IL-17R–Act1 complex, where it specifically phosphorylates Act1 at Ser311. This generates a docking site that recruits TRAF2 and TRAF5 but not TRAF6. In the absence of IL-17, SF2 (ASF) binds to the 3′ UTR of CXCL1 mRNA, causing its instability. After stimulation with IL-17, SF2 (ASF) associates with the TRAF2–TRAF5–Act1 complex, relieving the repression of CXCL1 mRNA and prolonging its half-life. Although this synergy requires the recruitment of Act1 to IL-17R, it occurs via TRAF6-independent stabilization of mRNA, which indicates the existence of an Act1-dependent, TRAF6-independent sig¬naling cascade separate from IL-17-induced activation of NF-κB. In addition, IL-17 activates mitogen-activated protein kinases in an Act1-dependent, TRAF6-independent manner, which further suggests that Act1 is an important bifurcation point for TRAF6-dependent and TRAF6-independent IL-17R signaling events (Ref.13).
IL-25, also known as IL-17E, is a high affinity ligand for IL-17RB. IL-17B is also reported to bind to IL-17RB with an affinity of 7.6 nM, while IL-25 binds to IL-17RB with Kd value of 1.1–1.4 nM. IL-25 appears to promote the expansion and further polarization of Th2 central memory cells .IL-17RA and IL-17RB as a signaling complex for IL-25 signaling.IL-17RA bound to the IL-17RB-IL-25 complex with an affinity of 14.1 ± 2.4 μM (Ref.14).Act1, the adaptor of IL-17RA signaling, is also involved in IL-17RB signaling. The interaction between Act1 and IL-17RB was abolished when the SEFIR domain was deleted in either Act1 or IL-17RB, indicating the recruitment of Act1 to IL-17RB is through the dimerization of the SEFIR domain.TRAF6 was implicated in IL-17RA signaling, NF-KappaB activation mediated through IL-17RB is abrogated in TRAF6 deficient cells, indicating IL-25 may acquire similar signaling pathways as IL-17. However, IL-25 activates ERK, JNK, and p38 by a TRAF6-independent mechanism. In contrary to IL-17RA that lacks TRAF6 binding sites, IL-17RB possesses TRAF6 binding sites. IL-17RB -mediated NF-B activation was attenuated in cells expressing in IL-17RB E338A, in which TRAF6-binding motif was mutated. IL-25 induces expression of GATA-3, c-maf, and JunB by activated Th2 memory cells. It is not known whether IL-25 mediates the signaling in CD4 T cells through TRAF6 and Act1 or there is independent pathway. In epithelial cells, IL-17RB is known to activate NF-KappaB and MAPK pathways through TRAF6 and TAK1. IL-17 has been associated with the pathogenesis of multiple autoimmune diseases including rheumatoid arthritis, multiple sclerosis and inflammatory bowel diseases. IL-17 is also important for host defence, further investigations into IL-17-mediated signaling to dissect the inflammatory pathway compared with the host defence pathway, and subsequent immunological and pathogenic roles of potential signaling targets, will help in the development of more specific medicines for alleviating symptoms associated with autoimmune inflammatory diseases without compromising host defence (Ref.1, 2&3).
Chang SH, Dong C.
Cell Signal. 2011 Jul;23(7):1069-75.2.IL-17/IL-17 receptor system in autoimmune disease: mechanisms and therapeutic potential.
Zhu S, Qian Y.
Clin Sci (Lond). 2012 Jun;122(11):487-511.3.IL-17 receptor signaling and T helper 17-mediated autoimmune demyelinating disease.
Zepp J, Wu L, Li X.
Trends Immunol. 2011 May; 32(5):232-9.
4.Structure and signalling in the IL-17 receptor family.
Gaffen SL.
Nat Rev Immunol. 2009 Aug; 9(8):556-67.
5.IL-17 and IL-22: siblings, not twins.
Eyerich S, Eyerich K, Cavani A, Schmidt-Weber C.
Trends Immunol. 2010 Sep;31(9):354-61.
6.IL-17RC: a partner in IL-17 signaling and beyond.
Ho AW, Gaffen SL.
Semin Immunopathol. 2010 Mar; 32(1):33-42.
7.IL-17F: regulation, signaling and function in inflammation.
Chang SH, Dong C.
Cytokine. 2009 Apr;46(1):7-11.
8.IL-17 activates the canonical NF-kappaB signaling pathway in autoimmune B cells of BXD2 mice to upregulate the expression of regulators of G-protein signaling 16.
Xie S, Li J, Wang JH, Wu Q, Yang P, Hsu HC, Smythies LE, Mountz JD.
J Immunol. 2010 Mar 1; 184(5):2289-96.
9.Differential regulation of the IL-17 receptor by gammac cytokines: inhibitory signaling by the phosphatidylinositol 3-kinase pathway.
Lindemann MJ, Hu Z, Benczik M, Liu KD, Gaffen SL.
J Biol Chem. 2008 May 16;283(20):14100-8. doi: 10.1074/jbc.M801357200.
10.IL-17 receptor signaling inhibits C/EBPbeta by sequential phosphorylation of the regulatory 2 domain.
Shen F, Li N, Gade P, Kalvakolanu DV, Weibley T, Doble B, Woodgett JR, Wood TD, Gaffen SL.
Sci Signal. 2009 Feb 24;2(59):ra8. doi: 10.1126/scisignal.2000066.
11.Structure-function relationships in the IL-17 receptor: implications for signal transduction and therapy.
Shen F, Gaffen SL.
Cytokine. 2008 Feb;41(2):92-104. Epub 2008 Jan 4.
12.TRAF6 dependent Act1 phosphorylation by the IKK-related kinases suppresses IL-17-induced NF-κB activation.
Qu F, Gao H, Zhu S, Shi P, Zhang Y, Liu Y, Jallal B, Yao Y, Shi Y, Qian Y.
Mol Cell Biol. 2012 Jul 30.
13.IL-17R signaling: new players get in on the Act1.
May MJ.
Nat Immunol. 2011 Aug 18; 12(9):813-5. doi: 10.1038/ni.2093.
14.A comparative study of ofloxacin and pivmecillinam in acute exacerbations of chronic bronchitis.
Egede F, Kristensen I.
Scand J Infect Dis Suppl. 1990; 68:46-9.
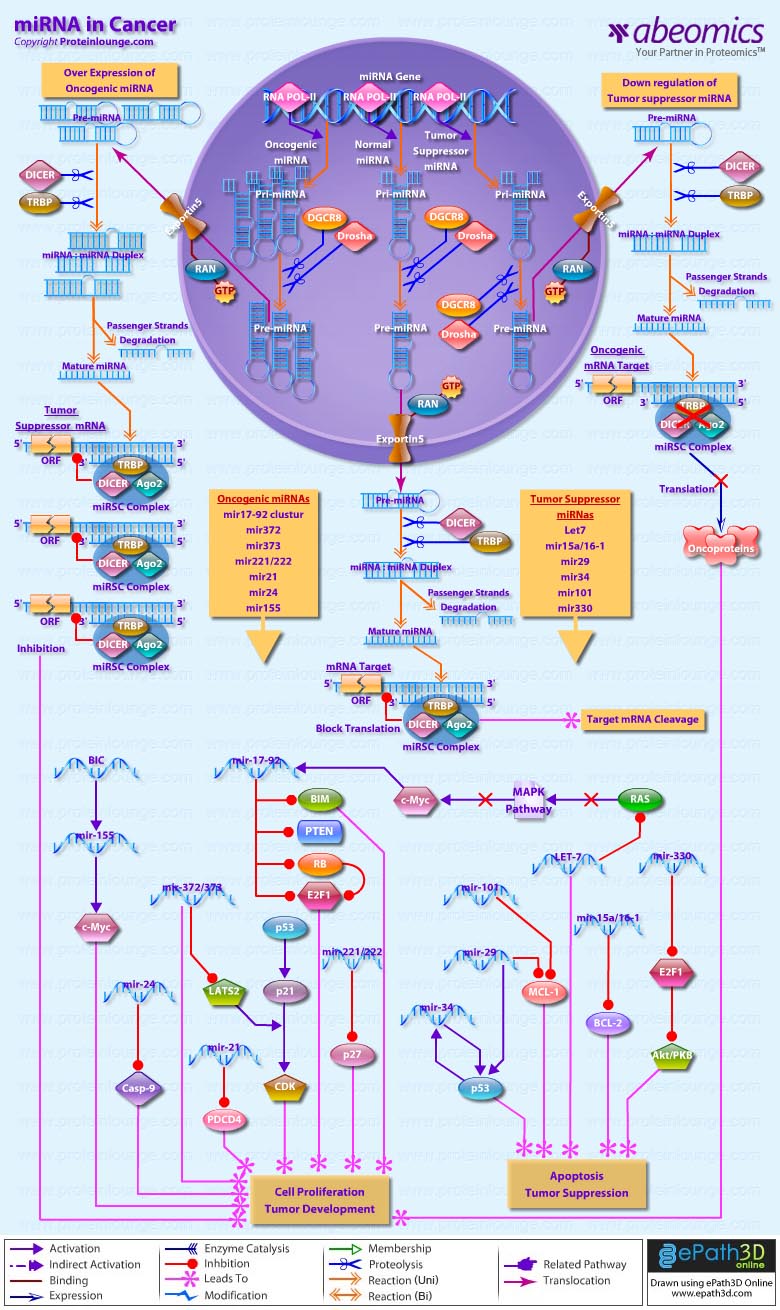
Over the last decade, a growing number of non-coding transcripts have been found to have roles in gene regulation and RNA processing. The most well known small non-coding RNAs are the miRNAs/miRs (microRNAs), which have been found to be involved in human tumorigenesis, revealing a new layer in the molecular architecture of cancer. Gene expression studies have shown that hundreds of miRNAs are deregulated in cancer cells, and functional studies have clarified that miRNAs are involved in all the molecular and biologic processes that drive tumorigenesis. miRNAs constitute a large class of phylogenetically conserved single-stranded RNA molecules of 19 to 25 nucleotides playing major role in post-transcriptional gene silencing by translational repression or mRNA cleavage (Ref. 1). Till date more than 500 miRNAs have been documented and the number is still growing. About 3% of human genes encode for miRNAs, and up to 30% of human protein coding genes are regulated by miRNAs (Ref. 2). Most miRNAs are transcribed by RNA Polymerase II as long pri-miRNAs (primary RNA) that contain a 5′ CAP structure and a 3’polyadenylated tail. The pri-miRNAs are processed in the nucleus by the RNase III enzyme, Drosha, and the double-stranded- RNA-binding protein, Pasha (also known as DGCR8), into ~70-nucleotide pre-miRNAs, which fold into imperfect stem-loop structures. The pre-miRNAs are then exported into the cytoplasm by the RAN-GTP dependent transporter exportin 5 and undergo an additional processing step in which a double- stranded RNA of ~22 nucleotides in length, referred to as the miRNA:miRNA* duplex, is excised from the pre-miRNA hairpin by another RNAse III enzyme, Dicer, in association with its RNA binding partner TRBP. Subsequently, the miRNA:miRNA* duplex is incorporated into the miRISC (multiprotein RNA-induced silencing) complex, which includes the Argonaute proteins (Argonate2), where the passenger strand is removed and degraded and subsequently the mature miRNA guides the RISC to target mRNAs whose expression can be inhibited by translational repression through perfect or nearly perfect complementarity to 3′ UTRs (Un-translated regions) or direct degradation of the target mRNA. It has been estimated that a single miRNA family can regulate as many as 200 different genes. Thus the effects of the miRNAs are pleotropic, and their aberrant expressions conceivably unbalance the cell¡¯s homeostasis, contributing to diseases, including cancer (Ref. 3 & 4).
miRNA expression correlates with various cancers, and these genes are thought to function as both tumor suppressors and oncogenes . About 50% of annotated human miRNAs are located in areas of the genome, known as fragile sites that are associated with cancer (Ref. 1). Expression of miRs and the role of miRs in cancer are tissue and tumor specific. Abnormal miR levels in tumors have important pathogenetic consequences: miRs may act as oncogenes or suppressor genes. miRs over expressed in tumors downregulate tumor suppressor genes, whereas miRs lost by tumors participate in oncogene overexpression (Ref. 5). In normal tissues, proper miRNA transcription, processing and binding to complementary sequences on the target mRNA results in the repression of target-gene expression through a block in protein translation or altered mRNA stability. The overall result is normal rates of cellular growth, proliferation, differentiation and cell death. The reduction or deletion of a miRNA that functions as a tumor suppressor leads to tumor formation. A reduction in or down regulation of mature miRNA levels can occur because of defects at any stage of miRNA biogenesis and ultimately leads to the inappropriate expression of the miRNA-target oncoprotein. The overall outcome might involve increased proliferation, invasiveness or angiogenesis, decreased levels of apoptosis, or undifferentiated or de-differentiated tissue, ultimately leading to tumor formation. The amplification or over expression of a miRNA that has an oncogenic role would also result in tumor formation. In this situation, increased amounts of a miRNA, which might be produced at inappropriate times or in the wrong tissues, would eliminate the expression of a miRNA-target tumor-suppressor gene and lead to cancer progression. Increased levels of mature miRNA might occur because of amplification of the miRNA gene, a constitutively active promoter, increased efficiency in miRNA processing or increased stability of the miRNA (Ref. 6).
Those miRNAs whose expression is increased in tumors are considered as oncogenes. These oncogene miRNAs, called ¡°oncomirs¡±, usually promote tumor development by negatively inhibiting tumor suppressor genes and/or genes that control cell differentiation or apoptosis (Ref. 7 & 6). mir-17¨C92 cluster is a classic example of an oncomir. This is a miRNA polycistron located at chromosome 13q31, a genomic locus that is amplified in lung cancer and several kinds of lymphoma, including diffuse large B-cell lymphoma. mir-17¨C92 cluster includes seven miRNAs: miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-20a, miR-19b-1 miR-92-1. Two tumor suppressor genes PTEN (phosphatase and tensin homolog deleted on chromosome ten) and RB2 are targeted by miR-17¨C92 cluster. PTEN promotes apoptosis through the P13K-Akt-PKB pathway. Over expression of mir-19 down regulates PTEN and leads to tumor development (Ref. 8, 9, 10 & 11). Unlike miR-19/PTEN axis toward apoptosis suppression, miR-17-92 increases proliferative capacity of Rb-deficient retinal cells. Deletion of Rb leads to compensatory up-regulation of the cyclin-dependent kinase inhibitor p21Cip1 and over expression of mir17-92 counteracts p21Cip1 up-regulation, promotes proliferation, and drive retinoblastoma formation. Over expression of mir-17¨C92 cluster also co-operates with c-Myc to accelerate lymphoma. Infect, miR-17-92 cluster itself is a transcriptional target of c-Myc. miR-17/20a targets E2F1, a regulator of the cell cycle and apoptosis. miR-92 targets BIM, a pro-apoptotic gene that counteracts the anti-apoptotic activity of genes such as Bcl2 (Ref. 9, 10 & 11). Another miRNA, miR-155, is linked with various lymphomas. This miRNA is encoded by a conserved region of the non-coding gene B- ell integration cluster, BIC, a common integration site for the avian leucosis virus, inducing B-cell lymphomas in collaboration with MYC (Ref. 3, 6 & 8). miR-372 and miR-373 are two additional examples of oncogenic miRNAs. These two miRNAs promote cell proliferation and tumor development by neutralizing p53-mediated CDK inhibition, possibly through direct inhibition of expression of the tumor suppressor gene LATS2 (Ref. 8 & 12). Another oncomir, mir-24, whose overexpression negatively regulates the proapoptotic factors caspase9 and APAF1, leading to cell proliferation and tumor development (Ref. 13 & 14). MiR-221 and miR-222 are two highly homologous microRNAs. These are positive regulators of prostate carcinoma growth through the repression of their target p27 which is a negative regulator of cell cycle progression (Ref. 15). In case of glioblastoma, over expression of oncogenic mir-21 inhibits its target programmed cell death 4 (Pdcd4), a known tumor-suppressor gene, and leads to increased proliferation of primary brain tumor cells (Ref. 16).
In oncogenesis, some miRNAs expression is decreased in cancerous cells. These types of miRNAs are considered tumor suppressor genes. Tumor suppressor miRNAs usually prevent tumor development by negatively inhibiting oncogenes and/or genes that control cell differentiation or apoptosis. Currently, several miRNAs are considered as tumor suppressor genes, for example, mir let-7, mir-15/16-1, mir-29, mir-34,mir-101& mir-330 (Ref. 8). let-7 is one of the founding members of the miRNA family. It is located at a chromosome region that is usually deleted in human cancers. let-7 is poorly expressed in lung cancers; reduced let-7 expression is significantly associated with shortened postoperative survival independent of disease stage. This confirms the tumor suppressor role of let-7. A recent study indicates that RAS oncogene is a direct target of let-7. let-7 negatively regulates RAS expression by pairing at the 3¡ä UTR of RAS mRNA for translational repression. Moreover let-7 negatively regulates the expression of RAS and MYC by targeting their mRNAs for translation repression (Ref. 8, 2 & 3). miRNAs encoded by the miR-15/16 cluster are also act as tumor suppressors. Expression of these miRNAs inhibits cell proliferation, promotes apoptosis of cancer cells, and suppresses tumorigenicity. Anti-apoptotic oncogene BCL2 is one of targets regulated by miR-15a and miR-16-1. Both these miRNAs negatively regulate BCL2 at a posttranscriptional level. Moreover, the Bcl2 repression by these miRNAs induces apoptosis in a chronic lymphocytic leukemia (Ref. 17 & 3). miR-101 and miR-29b exert their proapoptotic function via targeting Mcl-1,which is a member of Bcl-2 family and can inhibit apoptosis (Ref. 18 & 13).miR-29 also down-regulates p85a, a regulatory subunit of PI3K, and thereby enhances p53 activity through the negative loop between PI3K-AKT-MDM2 and p53 (Ref. 19). Another tumor suppressor miRNA, mir-34 promotes p53-mediated apoptosis, cell cycle arrest and senescence. Expression of miR-34 induces cell cycle arrest and thereby acts together with other effectors of the p53 tumor suppressor network to inhibit inappropriate cell proliferation. The expression of the mir-34 family members is regulated directly by p53, and consequently the expression of the mir-34 family reflects p53 activity (Ref. 2, 3, 19 & 13). microRNA mir-330 behaves like a potential tumor suppressor in many cancer types. It induces apoptosis of prostate cancer cells through E2F1- mediated suppression of Akt-phosphorylation (Ref. 18 & 20). Micro RNAs are causing tremendous excitement in cancer research. While the field of miRNAs and particularly the study of the roles of miRNAs in cancer is at an early stage, their potential as targeted therapeutic tools has not gone unnoticed. miRNA-based cancer therapies may be on the horizon in recent future.
Lee SK, Calin GA.
Discov Med. 2011 Mar;11(58):245-54.2.MicroRNA: implications for cancer.
Sassen S, Miska EA, Caldas C.
Virchows Arch. 2008 Jan;452(1):1-10.3.microRNAs and cancer: an overview.
Medina PP, Slack FJ.
Cell Cycle. 2008 Aug 15;7(16):2485-92.
4.The emerging roles of microRNAs in the molecular responses of metabolic rate depression.
Biggar KK, Storey KB.
J Mol Cell Biol. 2011 Jun;3(3):167-75.
5.MicroRNAs: a complex regulatory network drives the acquisition of malignant cell phenotype.
Santarpia L, Nicoloso M, Calin GA.
Endocr Relat Cancer. 2010 Jan 29;17(1):F51-75.
6.Oncomirs – microRNAs with a role in cancer.
Esquela-Kerscher A, Slack FJ.
Nat Rev Cancer. 2006 Apr;6(4):259-69.
7.Mechanisms and role of microRNA deregulation in cancer onset and progression.
Palmero EI, de Campos SG, Campos M, de Souza NC, Guerreiro ID, Carvalho AL, Marques MM.
Genet Mol Biol. 2011 Jul;34(3):363-70.
8.microRNAs as oncogenes and tumor suppressors.
Zhang B, Pan X, Cobb GP, Anderson TA.
Dev Biol. 2007 Feb 1;302(1):1-12.
9.Tumorigenicity of the miR-17-92 cluster distilled.
van Haaften G, Agami R.
Genes Dev. 2010 Jan 1;24(1):1-4.
10.miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma.
Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D.
Genes Dev. 2011 Aug 15;25(16):1734-45.
11.mir-17-92, a cluster of miRNAs in the midst of the cancer network.
Olive V, Jiang I, He L.
Int J Biochem Cell Biol. 2010 Aug;42(8):1348-54.
12.MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis.
Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q, Li YX, Liu M, Li X, Tang H.
J Biol Chem. 2011 Jul 22;286(29):25556-63.
13.MicroRNA: A matter of life or death.
Wang Z.
World J Biol Chem. 2010 Apr 26;1(4):41-54.
14.microRNA-24a is required to repress apoptosis in the developing neural retina.
Walker JC, Harland RM.
Genes Dev. 2009 May 1;23(9):1046-51.
15.The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice.
Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, Spagnoli LG, Farace MG, Ciafrè SA.
PLoS One. 2008;3(12):e4029.
16.Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo.
Gaur AB, Holbeck SL, Colburn NH, Israel MA.
Neuro Oncol. 2011 Jun;13(6):580-90.
17.miR-15a and miR-16-1 in cancer: discovery, function and future perspectives.
Aqeilan RI, Calin GA, Croce CM.
Cell Death Differ. 2010 Feb;17(2):215-20.
18.MicroRNAs in cardiac apoptosis.
Li P.
J Cardiovasc Transl Res. 2010 Jun;3(3):219-24.
19.Tumor suppressor p53 meets microRNAs.
Feng Z, Zhang C, Wu R, Hu W.
J Mol Cell Biol. 2011 Feb;3(1):44-50.
20.MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation.
Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, Lu PJ.
Oncogene. 2009 Sep 24;28(38):3360-70.
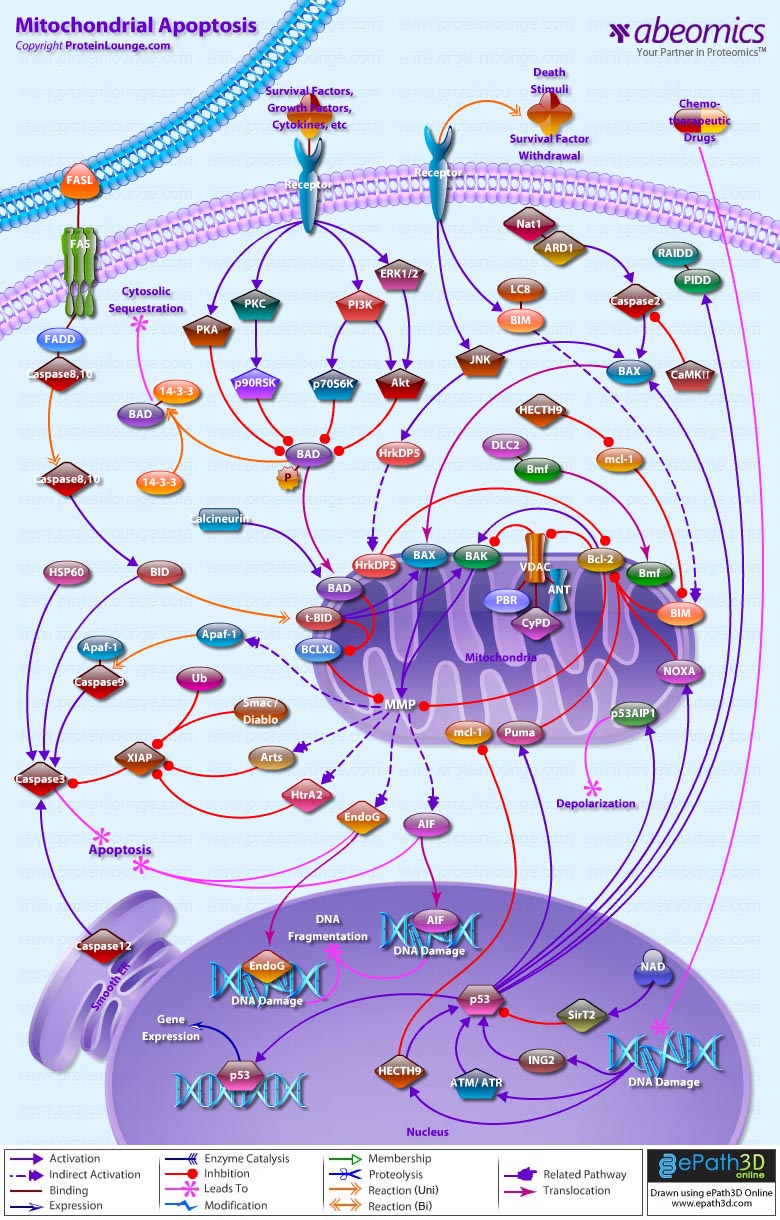
Apoptosis is a naturally occurring process by which a cell is directed to Programmed Cell Death. Apoptosis is based on a genetic program that is an indispensable part of the development and function of an organism. In this process, cells that are no longer needed or that will be detrimental to an organism or tissue are disposed of in a neat and orderly manner; this prevents the development of an inflammatory response, which is often associated with Necrotic cell death. There are at least two broad pathways that lead to Apoptosis, an “Extrinsic” and an “Intrinsic” Pathway. In both pathways, signaling results in the activation of a family of Cys (Cysteine) Proteases, named Caspases that act in a proteolytic cascade to dismantle and remove the dying cell. The extrinsic pathway begins outside a cell, when conditions in the extracellular environment determine that a cell must die. The intrinsic apoptosis pathway begins when an injury occurs within the cell. The injury could result in necrosis and produce an inflammatory response, but the apoptotic machinery is in place to ensure that the damaged cell is packaged and removed cleanly, in order to prevent inflammation. Apoptosis in Mitochondria is the best known intrinsic apoptosis pathway (Ref.1 & 2).
The Mitochondrial pathway of apoptosis functions in response to various types of intracellular stress including Growth Factor withdrawal, DNA damage, unfolding stresses in the Endoplasmic reticulum and Death Receptor stimulation. Fas (CD95/Apo1), a member of the TNFR family, typify the classical view of DR (Death Receptor) function and play an important role in stimulating Mitochondrial apoptotic pathway. FasL (Fas Ligand), a homotrimeric protein acts as ligand for Fas and causes oligomerization of its Receptor on binding. Associated with this is the clustering of the Death Domains and binding of cofactor FADD (Fas-Associated via Death Domain). The FADD protein binds via its DED (Death Effector Domain) motif to a homologous motif in Procaspase8. The complex of Fas, FADD and ProCaspase8 is called the DISC (Death Inducing Signaling Complex). The cofactor function of FADD, in turn, is blocked by interaction with the regulator FLIP (FLICE Inhibitory Protein). Upon recruitment by FADD, Procaspase8 oligomerization drives its activation through self-cleavage. Active Caspase8 then activates downstream Caspases (Caspase3 and 7). Activated Caspase8 activates Caspase3 through two pathways. In the first pathway Caspase8 cleaves BID (Bcl2 Interacting Protein) and its COOH-terminal part translocates to Mitochondria where it triggers CytoC (Cytochrome-C) release. The released CytoC binds to APAF1 (Apoptotic Protease Activating Factor-1) together with dATP and Procaspase9 and activates Caspase9. The Caspase9 cleaves Procaspase3 and activates Caspase3. Another pathway is that Caspase8 cleaves Procaspase3 directly and activates it. Caspase 3 then commits the cell to Apoptosis. Besides DRs, Growth Factors also influence Mitochondrial Apoptosis via PI3K (Phosphatidylinositde-3 Kinase) and Akt (v-Akt Murine Thymoma Viral Oncogene Homolog) pathways. Growth Factors bind to Growth factor receptors and activate PI3K. Activation of PI3K pathways leads to Akt activation. Akt is very important in BAD (Bcl2-Antagonist of Cell Death) regulation, which is a proapoptotic member of Bcl2 (B-Cell Leukemia-2) family and is involved in Mitochondrial Apoptosis (Ref.6). PKC (Protein Kinase-C) may also play an important role in Apoptosis by activating p90RSKs (Ribosomal-S6 Kinases), which inhibits BAD (Ref. 3, 4 & 5).
In addition to Receptor-mediated Apoptosis, there is another pathway activated by various forms of Cellular Stress. Intrinsic stresses such as Oncogenes, direct DNA damage, Hypoxia, and Survival factor deprivation, can activate the Intrinsic Apoptotic pathway. p53 is a sensor of cellular stress and is a critical activator of the intrinsic pathway. The DNA checkpoints proteins, ATM (Ataxia Telangiectasia Mutated protein), and Chk2 (Checkpoints Factor-2) directly phosphorylate and stabilize p53 and inhibit MDM2 (Mouse Double Minute-2 Homolog) mediated ubiquitination of p53. MDM2 binds p53 and mediates the nuclear export. When bound to MDM2, p53 can no longer function as an activator of transcription. p53 initiates Apoptosis by transcriptionally activating proapoptotic Bcl2 family members and repressing antiapoptotic Bcl2 proteins and CIAPs (Cellular Inhibitor of Apoptosis). Other p53 targets include BAX (BCL2 Associated X-protein), Noxa, PUMA (p53-Upregulated Modulator of Apoptosis) and the most recently identified, BID. p53 also transactivates other genes that may contribute to Apoptosis including PTEN (Phosphatase and Tensin Homolog Deleted On Chromosome-10), APAF1, Perp, p53AIP1 (p53-regulated Apoptosis-Inducing Protein-1), and genes that lead to increases in ROS (Reactive Oxygen Species). These ROS lead to generalized oxidative damage to all Mitochondrial components. Damage to Mitochondrial DNA disrupts Mitochondrial oxidative phosphorylation, contributing to a number of Human diseases (Ref. 5 & 6).
Following the reception of stress signals, proapoptotic Bcl2 family proteins are activated and subsequently interact with and inactivate antiapoptotic Bcl2 proteins. Bcl2 family consists of both anti- and proapoptotic members that possess conserved alpha-helices with sequence conservation clustered in Bcl2 Homlogy domains. Proapoptotic members include “Multidomain” BAX family proteins such as BAX, BAK (Bcl2 Antagonist Killer) etc. that display sequence conservation in BH1-3 region and “BH3-only” proteins such as BID, BAD, Noxa, PUMA and BIM (BCL2-Interacting Protein) that have only the short BH3 motif. BAX family proteins act downstream in Mitochondrial disruption, whereas BH3-only proteins act upstream in the pathway, detecting developmental death cures or intracellular damages. Anti-apoptotic members like Bcl2, BclXL and their relatives exhibit homology in all segments BH1-4. The antiapoptotic and proapoptotic protein interaction leads to the destabilization of the Mitochondrial membrane and release of apoptotic factors including CytoC, SMAC (Second Mitochondria-Derived Activator of Caspase), Arts and Omi/HTRA2 (High Temperature Requirement Protein-A2). Bcl2L (Bcl2-Like), BCLXL and other anti-apoptotic BCL2 family members reside in the outer Mitochondrial membrane and prevent CytoC release. BAX , BID and BIM are initially inactive and must translocate to Mitochondria to induce apoptosis, either by forming pores in Mitochondria directly or by binding via BH3 domains to BCL2, BCLXL and BFL1, and antagonizing these anti-apoptotic proteins. MMP (Mitochondrial Membrane Permeabilization) is clearly a pivotal event in the progression of Apoptosis in many systems. At least two mechanisms of MMP have been described. First mechanism proposes that VDAC (Voltage-Dependent Anion Channel), ANT (Adenine Nucleotide Transporter), PBR (Peripheral-type Benzodiazepine Receptor) and CypD (Cyclophilin-D) come together to form the PTPC (Permeability Transition Pore Complex). This pore complex may also associate with BAX, BAK1 or BIM, which accelerate channel opening or BclXL, which causes closure. According to the second mechanism BAX is released from its interaction with 14-3-3 and translocates from cytosol to Mitochondria in response to diverse signals. Here it oligomerizes forming protein pores. Pore formation also takes place in conjunction with the BH3-only Bcl-2 family member BID upon proteolysis to tBID (Truncated BID). BAK1 is located at Mitochondria and has similar pore forming properties to BAX, via its oligomerization and association with tBID. BID is activated by Caspase8-induced cleavage during DR signaling, whereas BIM is released from its association with microtubules (Ref. 5 & 7).
Other proteins released from Damaged Mitochondria, SMAC (Second Mitochondria-Derived Activator of Caspase)/ Diablo, Arts and Omi/HTRA2 (High Temperature Requirement Protein-A2), counteract the effect of IAPs (Inhibitor of Apoptosis Proteins), which normally bind and prevent activation of Caspase3. The interaction between Bcl family members, IAPs, SMAC and Omi/HTRA2 is central to the intrinsic Apoptosis pathway. Recent studies demonstrated that another nuclease, EndoG (Endonuclease-G), is specifically activated by Apoptotic stimuli and is able to induce nucleosomal fragmentation of DNA independently of Caspase and DFF (DNA-Fragmentation Factor)/ CAD (Caspase-Activated DNAse). EndoG is a mitochondrion-specific nuclease that translocates to the nucleus and cleaves chromatin DNA during Apoptosis. Another protein, AIF (Apoptosis Inducing Factor) has also been attributed a role in Apoptosis, becoming active upon translocation from Mitochondria to nuclei, where it initiates chromatin condensation and large-scale DNA fragmentation (Ref.9). Endoplasmic reticulum stress also takes part in apoptosis by activating Caspase3 via Caspase12. Programmed cell death and its morphologic manifestation of Apoptosis is a conserved pathway that in its basic tenets appears operative in all metazoans. Apoptosis also operates in adult organisms to maintain normal cellular homeostasis. This is especially critical in long-lived mammals that must integrate multiple physiological as well as pathological death signals, which for example includes regulating the response to infectious agents. Gain and loss of function models of genes in the core Apoptotic pathway indicate that the violation of cellular homeostasis can be a primary pathogenic event that results in disease. Evidence indicates that insufficient Apoptosis can manifest as Cancer or Autoimmunity, while accelerated cell death is evident in Acute and Chronic Degenerative diseases, Immunodeficiency, and Infertility (Ref. 8 & 9).
Wilk S.
Sci STKE. 2005 May 24;2005(285):tr16.2.Apoptosome formation and caspase activation: is it different in the heart?
Czerski L, Nunez G.
J Mol Cell Cardiol. 2004 Sep;37(3):643-52. Review.3.Role of mitochondria as the gardens of cell death.
Kim R, Emi M, Tanabe K
Cancer Chemother Pharmacol. 2005 Sep 21;:1-9
4.Cutting edge: innate immunity conferred by B cells is regulated by caspase-8.
Beisner DR, Ch’en IL, Kolla RV, Hoffmann A, Hedrick SM.
J Immunol. 2005 Sep 15;175(6):3469-73.
5.TRAIL-induced apoptosis in human vascular endothelium is regulated by phosphatidylinositol 3-kinase/Akt through the short form of cellular FLIP and Bcl-2.
Alladina SJ, Song JH, Davidge ST, Hao C, Easton AS.
J Vasc Res. 2005 Jul-Aug;42(4):337-47. Epub 2005 Jun 28.
6.The p53 pathway: positive and negative feedback loops.
Harris SL, Levine AJ.
Oncogene. 2005 Apr 18;24(17):2899-908. Review.
7.Unknotting the roles of Bcl-2 and Bcl-xL in cell death.
Kim R.
Biochem Biophys Res Commun. 2005 Jul 29;333(2):336-43.
8.Endoplasmic reticulum quality control and apoptosis.
Groenendyk J, Michalak M.
Acta Biochim Pol. 2005;52(2):381-95. Epub 2005 May 31.
9.Mitochondrial dysfunction and heart disease.
Rosenberg P.
Mitochondrion. 2004 Sep;4(5-6):621-8. Epub 2004 Oct 28.
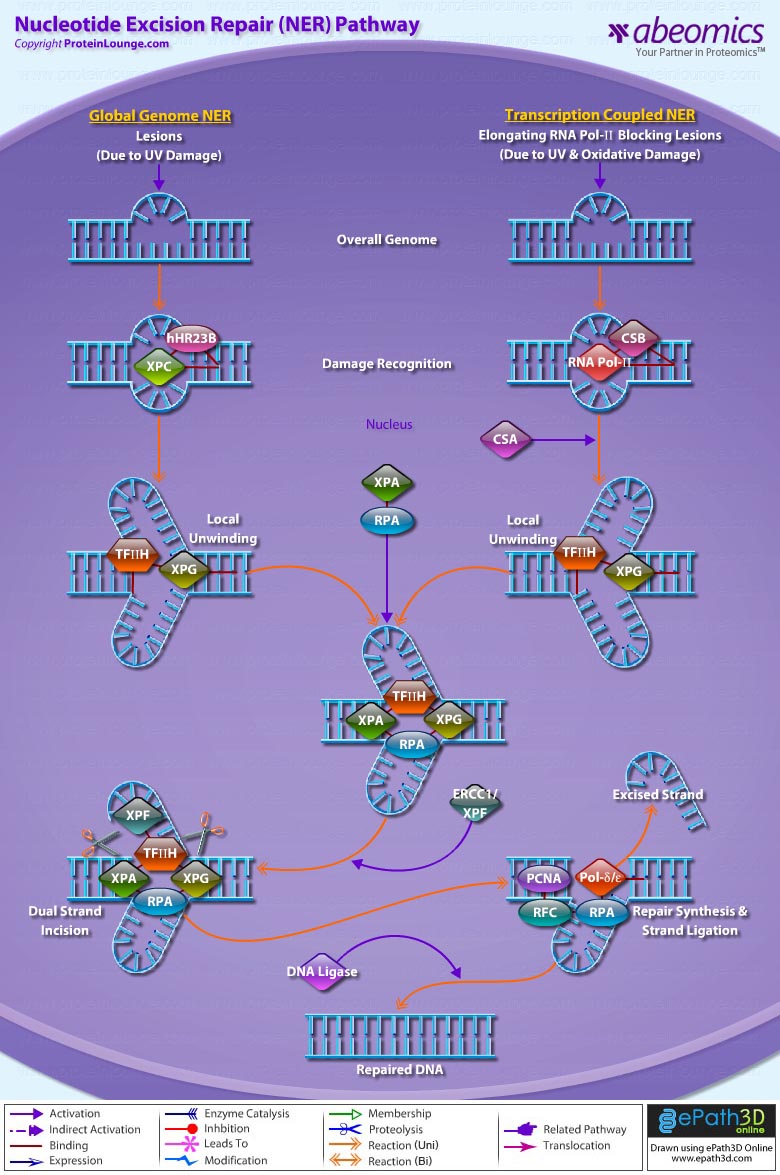
The inability to repair DNA damage properly leads to various disorders and enhanced rates of tumor development. Mammals respond to chromosomal insults by activating a complex damage response pathway. These pathways regulate known responses such as cell cycle arrest and apoptosis, and have recently been shown to control additional processes including direct activation of DNA repair networks (Ref.1). Different DNA-repair pathways operate on different types of DNA lesions. NER (Nucleotide Excision Repair) is the most flexible of the DNA repair pathways considering the diversity of DNA lesions it acts upon. The most significant of these lesions are pyrimidine dimers caused by the UV component of sunlight. Other NER substrates include bulky chemical adducts, DNA intrastrand cross links, and some forms of oxidative damage. The common features of lesions recognized by the NER pathway are that they cause both a helical distortion of the DNA duplex and a modification of the DNA chemistry (Ref.2).
The NER process requires the action of more than 30 proteins in a stepwise manner that includes damage recognition, local opening of the DNA duplex around the lesion, dual incision of the damaged DNA strand, gap repair synthesis, and strand ligation. There are two distinct forms of NER: GG-NER (Global Genomic NER), which corrects damage in transcriptionally silent areas of the genome, and TC-NER (Transcription Coupled NER), which repairs lesions on the actively transcribed strand of the DNA. These two subpathways are fundamentally identical except in their mechanism of damage recognition. In GG-NER, the XPC (Xeroderma Pigmentosum, Complementation Group C) protein complex is responsible for the initial detection of damaged DNA (Ref.5). Conversely, damage recognition during TC-NER does not require XPC, but occurs when the transcription machinery is stalled at the site of injury. The stalled RNA Polymerase complex is displaced in order to allow the NER proteins to access the damaged DNA. This displacement is aided by the action of the CSA (Cockayne Syndrome A) and CSB proteins, as well as other TC-NER-specific factors. The subsequent steps of GG- and TC-NER proceed in an essentially identical manner. XPA and the heterotrimeric RPA (Replication Protein A) then bind at the site of injury and further aid in damage recognition. Next, the XPB and XPD helicases, components of the multi-subunit Transcription Factor, TFIIH, unwind the DNA duplex in the immediate vicinity of the lesion for the enzyme RNA Polymerase-II to begin transcription. The Endonucleases XPG and ERCC1 (Excision Repair Cross-Complementing Group-1)/XPF then cleave one strand of the DNA at positions 3′ and 5′ to the damage, respectively, generating an approximately 30 base oligonucleotide containing the lesion. This oligonucleotide is displaced, making way for gap repair synthesis (performed by DNA Pol Delta/Epsilon, as well as several replication accessory factors). Finally the nick in the repaired strand is sealed by a DNA Ligase, thus completing the NER process (Ref.3).
NER deals with the wide class of helix-distorting lesions that interfere with base pairing and generally obstruct transcription and normal replication. Most NER lesions arise from exogenous sources (except for some oxidative lesions) (Ref.4). At least three syndromes are associated with inborn defects in NER: XP, CS and TTD (Trichothiodystrophy), all characterized by exquisite sun sensitivity (Ref.5). The prototype repair disorder, XP exhibits a dramatic >1000-fold incidence of sun-induced skin cancer (basal cell carcinoma, squamous cell carcinoma and melanoma). The disorder arises from mutations in one of seven genes (XPA–XPG) (Ref.6). CS, caused by mutation in the CSA or CSB genes, is a TC-NER specific disorder that is remarkably dissimilar from XP. No predisposition to cancer is observed because TC-NER defect causes CS cells to be particularly sensitive to lesion-induced apoptosis, thereby protecting against tumorigenesis. Physical and neurological developments are impaired, resulting in dwarfism and dysmyelination. The syndrome includes features of premature ageing, which is related to the increased trigger for apoptosis induced by transcriptional arrest from endogenous lesions in combination with the TC-NER defect. TTD is a condition sharing many symptoms with CS, but with the additional hallmarks of brittle hair, nails and scaly skin. TTD is not associated with cancer. Mutations in the XPD or XPB genes give rise to all three diseases. Thus, as subunits of TFIIH, XPB and XPD have dual functions: NER and transcription initiation. Mutations not only compromise NER, but also affect transcription, causing developmental delay and reduced expression of the matrix proteins that causes brittle hair and scaly skin (Ref.4).
Zhou BB, Elledge SJ.
Nature. 2000 Nov 23; 408(6811): 433-9. Review.2.Bipartite substrate discrimination by human nucleotide excision repair.
Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H.
Proc Natl Acad Sci U S A. 1997 Jun 24; 94(13): 6664-9.3.DNA repair. The bases for Cockayne syndrome.
Hanawalt PC.
Nature. 2000 May 25; 405(6785): 415-6. No abstract available.
4.Genome maintenance mechanisms for preventing cancer.
Hoeijmakers JH.
Nature. 2001 May 17; 411(6835): 366-74. Review.
5.The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases.
Lehmann AR.
Genes Dev. 2001 Jan 1; 15(1): 15-23. Review. No abstract available.
6.DNA repair. Variants on a theme.
Wood RD.
Nature. 1999 Jun 17; 399(6737): 639-40. No abstract available.
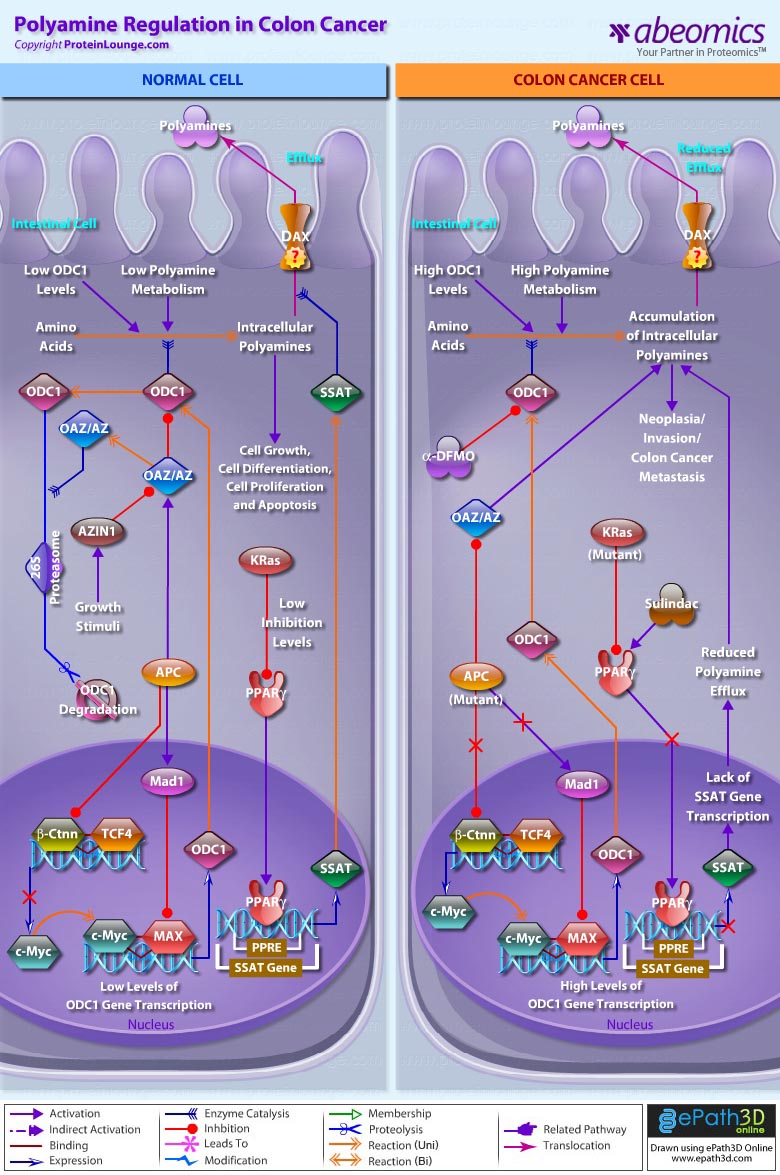
Polyamines are vital for the growth and function of normal cells. The complexity of polyamine metabolism and the multitude of compensatory mechanisms that are invoked to maintain polyamine homoeostasis argue that these amines are critical to cell survival. The regulation of polyamine content within cells occurs at several levels, including transcription and translation (Ref.1). The amino-acid derived polyamines like Putrescine, Spermidine and Spermine that are the main polyamines found in prokaryotes and eukaryotes, have long been associated with cell growth and cancer, and specific oncogenes and tumor suppressor genes regulate polyamine metabolism. Regulation of these polyamines (otherwise known as “organic cations”) is chiefly associated with cancers pertaining to the colon. They are formed by the enzymatic decarboxylation of the amino acids like Ornithine or Arginine by ODC1 (Ornithine Decarboxylase-1) the enzyme required for the first stage in polyamine synthesis. ODC1 is subject to both positive and negative feedback regulation by polyamines: high polyamine concentrations decrease, and low polyamine concentrations increase, activity (Ref.2 & 3).
In normal cells, low ODC1 and polyamines level is essential for normal cell growth and differentiation, cell proliferation and apoptosis, for which, the feedback regulation of ODC1, is a mixture of post-transcriptional regulation and the induction of a unique ODC1-specific inhibitor termed OAZ (Ornithine Decarboxylase Antizyme). OAZ binds to ODC1 and the OAZ-ODC1 complex is degraded by the 26S Proteasome. Unusually, the degradation of ODC1 by this Proteasome occurs in an ATP-dependent, but Ub (Ubiquitin)-independent, manner. ODC1 is released from OAZ by another unique protein, called AZIN1 (Antizyme Inhibitor-1), which liberates ODC1 in the presence of growth stimuli by virtue of having a higher affinity for OAZ than for ODC1. Additionally, OAZ can alter polyamine homoeostasis by down-regulating polyamine uptake independent of the effects on ODC1. It may be that OAZ binding to ODC1 causes a conformational change in ODC1, resulting in exposure of its C-terminus, so targeting it for degradation. Also expression of full-length APC (Adenomatous Polyposis Coli) results in up-regulation of OAZ and APC-dependent expression of Mad1 forms hetero-complexes with the c-Myc binding partner, MAX (Myc-Associated Factor-X). c-Myc acts as a transcriptional activator and Mad1 as the transcriptional repressor for ODC1 gene expression. Mad1 compete with c-Myc for binding to MAX, thereby decreasing the levels of c-Myc:MAX complexes. An increased ratio of Mad1:MAX to c-Myc:MAX hetero-complexes lead to greater binding at e-box domains by Mad1:MAX complexes. So the expression of c-Myc is down-regulated during differentiation, whereas Mad1 expression is up-regulated and the reciprocal effect results in cell growth by maintaing low levels of ODC1 and polyamines. APC further inhibits Ctnn-Beta (Catenin-Beta)/TCF4 (Transcription Factor-4) association to down-regulate ODC1 gene expression (Ref.1 & 4). In addition, SSAT (Spermidine/SpermineN1-Acetyltransferase) gene transcription is negatively regulated by the KRas (Kirsten-Ras) oncogene through a mechanism involving PPAR-Gamma (Peroxisome Proliferative Activated Receptor-Gamma), a putative tumor suppressor. PPAR-Gamma positively regulates the transcription of SSAT through PPRE (PPAR Response Element) in the SSAT promoter. KRas suppresses SSAT transcription by inhibiting PPAR-Gamma expression and subsequent binding to the SSAT promoter. This maintains polyamine homeostasis and regulates polyamine efflux through DAX (Diamine Exporter). The mechanisms of this polyamine export, or efflux from mammalian cell types, are not yet defined (Ref.3).
In contrast, when wild-type APC is lost in intestinal epithelia, during development of colon cancer, decreased OAZ activity contributes to increased ODC levels. Defects in the APC gene may occur in at least 90% of colon adenomas and cancers and are responsible for both inherited and sporadic forms. However, the mechanisms by which APC mutations promote tumorigenesis are poorly understood (Ref.4). It is known that tumor cells with truncated APC exhibit high levels of Ctnn-Beta because it cannot be targeted to the 26S Proteasome for degradation. This leads to Ctnn-Beta/TCF4 association and translocation to the nucleus where transcription of target gene, c-Myc is activated, leading to high levels of ODC and intracellular polyamine accumulation. Similarly, KRas which is commonly mutated and aberrantly activated in human colon cancer and other gastrointestinal cancers strongly inhibits PPAR-Gamma expression and subsequent binding to the SSAT promoter. This results in loss of polyamine efflux and facilitates neoplasia/invasion by cancer cells. So, polyamine synthesis and catabolism are both regulated by signaling pathways that are influenced by oncogenes and tumour-suppressor genes. Aberrant APC and KRas expression are essential for neoplasia indicate how polyamine levels become increased in colon and other gastrointestinal cancers (Ref.5 & 6).
Polyamines continue to be molecules that hold fascination for biologists, chemists, molecular biologists and clinical researchers. Polyamine levels can increase because of synthesis and/or uptake, and their metabolism is highly regulated by a series of catabolic enzymes in mammals. Consequently, it should not be surprising that attaining the goal of reducing high polyamine levels in tissues during cancer development might require targeting two or more of these processes. The concept of combination therapy for chemoprevention is important, as combinations of agents are generally more effective than single agents. Although anticancer agents such as Alpha-DFMO (Alpha-Difluoromethylornithine) (inhibit ODC1) and Sulindac (activates PPAR-Gamma) function, are effective in treating colon cancer yet the future may witness adoption of new functional genomics approaches like RNAi technology that provides the opportunity to pursue an exciting new therapeutic approach to treat colon cancers (Ref.7 & 8).
Wallace HM, Fraser AV, Hughes A.
Biochem. J. 2003 Nov 15;376(Pt 1):1-14.2.Polyamines and cancer: old molecules, new understanding.
Gerner EW, Meyskens FL Jr.
Nat. Rev. Cancer. 2004 Oct;4(10):781-92.3.Signaling pathways in intestinal development and cancer.
Sancho E, Batlle E, Clevers H.
Annu. Rev. Cell Dev. Biol. 2004;20:695-723.
4.APC-dependent regulation of ornithine decarboxylase in human colon tumor cells.
Fultz KE, Gerner EW.
Mol. Carcinog. 2002 May;34(1):10-8.
5.Polyamines as modifiers of genetic risk factors in human intestinal cancers.
Babbar N, Gerner EW.
Biochem. Soc. Trans. 2003 Apr;31(2):388-92.
6.Polyamines and colon cancer.
Milovic V, Turchanowa L.
Biochem. Soc. Trans. 2003 Apr;31(2):381-3.
7.RNAi technology: a revolutionary tool for the colorectal cancer therapeutics.
Lv W, Zhang C, Hao J.
World J. Gastroenterol. 2006 Aug 7;12(29):4636-9.
8.Cyclooxygenase-independent induction of apoptosis by sulindac sulfone is mediated by polyamines in colon cancer.
Babbar N, Ignatenko NA, Casero RA Jr, Gerner EW.
J. Biol. Chem. 2003 Nov 28;278(48):47762-75
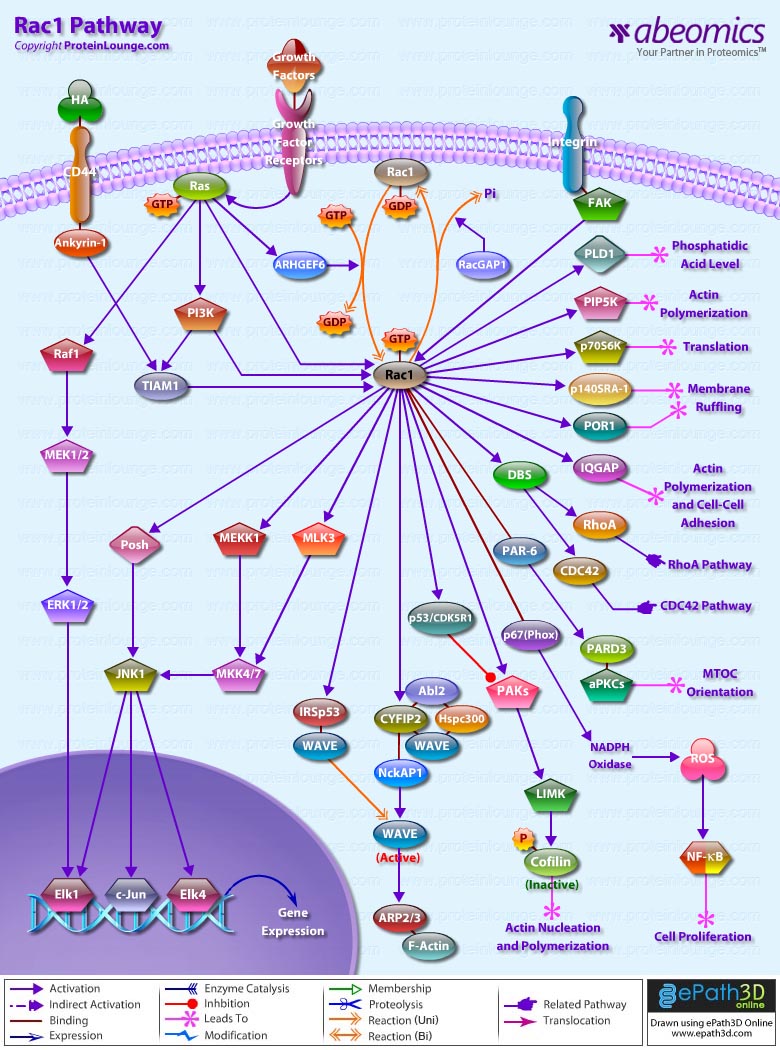
To achieve strong adhesion to their neighbors and sustain stress and tension, epithelial cells develop many different specialized adhesive structures. Breakdown of these structures occurs during tumor progression with the development of a fibroblastic morphology characteristic of metastatic cells. Adhesion receptors of the Cadherin family have been implicated in these cellular processes, which play an important role in the development and maintenance of the differentiated epithelial phenotype during organogenesis and adult life. Cadherin-mediated adhesion requires the activity of the cytosolic proteins of the Rho subfamily members, Rho, Rac and CDC42 (Cell Division Cycle-42). They belong to the Ras Superfamily of small GTPases, whose function is regulated depending on the type of guanine nucleotide bound. Mammals have three closely related Rac isoforms, Rac1, Rac2 and Rac3 (Ref.1).
Rac1 is a small G-protein in the Rho family that drives Actin polymerization and formation of lamellipodia, promotes cell-cell adhesion, breakdown and migration of different carcinoma cells. In other epithelial cell lines, Rac activation also plays a role in scattering after distinct stimuli such as Growth factor stimulation or Integrin engagement. It also plays a critical role in processes, such as control of cell morphology, transcriptional activation, and apoptosis signaling. The broad range of events controlled by this GTPase requires regulation of its interactions with multiple downstream targets. Rac1 is activated by GEF (Guanine Nucleotide Exchange Factors), in particular ARHGEF6 and repressed by GAPs (GTPase-Activating Proteins), specifically RacGAP. Rac1 GTPase mediates key cellular processes in response to upstream regulators such as Growth Factors, Integrins and HA (Hyaluronic Acid)-binding receptor CD44. Growth Factors and Growth Factor Receptors stimulate Ras by recruiting SOS (Son of Sevenless), GRB2 (Growth Factor Receptor-Bound Protein-2) and SHC ({SHC (Src Homology-2 Domain Containing) Transforming Protein}) to the membrane in order to activate PI3K (Phosphatidylinositde-3 Kinase) or phosphorylate Src to recruit PI3K. Rac is a key downstream target/effector of PI3K. Rac1 can also be activated by Integrin via FAK (Focal Adhesion Kinase). Interaction between CD44 (the HA-binding Receptor) and TIAM1 (T-lymphoma invasion and metastasis 1) can also activate Rac1. TIAM1 is one of the known GDP/GTP exchange factors for Rac1. TIAM1 and the cytoskeletal protein, Ankyrin, physically associate as a complex in vivo. In particular, the ARD (Ankyrin Repeat Domain) of Ankyrin is responsible for TIAM1 binding. Ankyrin binding to TIAM1 activates Rac1. Upon activation, Rac1 interacts with and regulate a spectrum of functionally diverse downstream effectors, initiating a network of cytoplasmic and nuclear signaling cascades (Ref.2, 3 & 4).
A number of proteins act as targets for Rac1 including PAKs (p21-Activated Kinases), IQGAP1 (IQ motif containing GTPase Activating Protein-1), MRCK/ CDC42BPA (CDC42 binding protein kinase Alpha (DMPK-like), POR1 (Partner Of Rac1) and POSH (Plenty Of SH3s). Rac binds p67(Phox) to increase activation of the NADPH Oxidase system and production of ROS (Reactive Oxygen Species), which mediate activation of NF-KappaB (Nuclear Factor-KappaB)-dependent gene expression, effects of Rac on cell cycle progression and inhibition of Rho activity. Rac binds the WAVE (WASP Family Verprolin Homology Domain-Containing Protein) complex (also containing Abi and IRSp53/58), to release active WAVE, which promotes actin polymerization in lamellipodia through activation of the ARP2/3 (Actin-Related Protein-2/3) complex. Rac also binds to the actin-binding protein IQGAP, which is implicated in regulation of cell-cell adhesion and Microtubule orientation. By binding to the microtubule tip protein Clip170, IQGAP1 captures growing microtubules at the leading edge of migrating fibroblasts, which results in cell polarization. Recently, a novel Rac-interacting protein, POR1 (Partner of Rac1), has been shown to play a role in membrane ruffling. p140SRA1 (Specifically Rac1-Associated Protein) is also a novel Specific target for Rac1 Small GTPase and is also involved in membrane ruffling. Recent findings Rac, involved in the dynamics of Actin cytoskeleton and cell polarity, bind to a protein complex containing PAR-6, PAR-3/ASIP, and aPKC (atypical Protein Kinase-C) (Ref.1, 5 & 6).
Members of the Rho subfamily of GTP-binding proteins including Rho and Rac are also implicated in the regulation of PLD (Phospholipase-D). PLD catalyzes the hydrolysis of Phosphatidylcholine to yield Phosphatidic Acid and Choline. Phosphatidic Acid is a second messenger involved in membrane remodeling events that are critical to cell growth, such as vesicle trafficking and regulated secretion. Both Rac and Rho bind to and activate PIP5K (Phosphatidylinositol-4-Phosphate 5-Kinase), which increases the amount of PIP2 (Phosphatidylinositol (4,5)-bisphosphate). Rac1 coordinately activates p70S6K (p70 Ribosomal -S6 Kinase) and JNK (c-Jun Kinase) via MLK3 (Mixed-Lineage Protein Kinase-3) activation. MLK3 activates JNK via MKK4/7 (MAP Kinase Kinase-4/7). JNK, once activated enter the nucleus and phosphorylate transcription factors such as c-Jun, c-Fos, Elk1 and Elk4. Rac also activates DBS (Dbl’s Big Sister), which further activates RhoA and CDC42. In neurons, Rac acts through the protein kinase CDK5 (Cyclin-Dependent Kinase-5) and p35 to phosphorylate and downregulate PAK1, increasing neuronal migration. PAK1 also phosphorylates and activates LIMK (LIM Kinase), which phosphorylates Cofilin as one target. Cofilin stimulates actin depolymerization and changes in cell structure, and phosphorylation of Cofilin by LIMK represses its activity (Ref.7, 8 & 9).
Signaling pathways that are regulated by Rho family members play an important role in several pathological conditions, including cancer, inflammation, and bacterial infections. Rac controls the generation of ROS , both in leukocytes and nonhematopoietic cells and is necessary for Cadherin-dependent adhesion. Rac activation is required for the full transformed phenotype induced by oncogenes such as TIAM1 and Ras. In addition, Rac activation perturbs cadherin contacts with a concomitant change in cell shape, including formation of lamellae/protusions and conversion to a fibroblastic morphology. Translocation of Rac to the plasma membrane is also required for assembly and activation of the NADPH Oxidase complex (Ref.2 & 10).
Kimura K, Kawamoto K, Teranishi S, Nishida T.
Invest Ophthalmol Vis Sci. 2006 Oct;47(10):4323-9.2.Rac1 and Rac2 regulate macrophage morphology but are not essential for migration.
Wheeler AP, Wells CM, Smith SD, Vega FM, Henderson RB, Tybulewicz VL, Ridley AJ.
J Cell Sci. 2006 Jul 1;119(Pt 13):2749-57. Epub 2006 Jun 13.3.Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration.
Murai T, Miyazaki Y, Nishinakamura H, Sugahara KN, Miyauchi T, Sako Y, Yanagida T, Miyasaka M.
J Biol Chem. 2004 Feb 6;279(6):4541-50. Epub 2003 Nov 17.
4.Expressions of Rac1, Tiam1 and Cdc42 in retinoblastoma.
Adithi M, Venkatesan N, Kandalam M, Biswas J, Krishnakumar S.
Exp Eye Res. 2006 Oct 4;
5.WAVE signalling: from biochemistry to biology.
Soderling SH, Scott JD.
Biochem Soc Trans. 2006 Feb;34(Pt 1):73-6.
6.Rac-1 and IQGAP are potential regulators of E-cadherin-catenin interactions during murine preimplantation development.
Natale DR, Watson AJ.
Mech Dev. 2002 Dec;119 Suppl 1:S21-6.
7.Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D.
Santy LC, Casanova JE.J Cell Biol. 2001 Aug 6;154(3):599-610. Epub 2001 Jul 30.
8.RhoG signals in parallel with Rac1 and Cdc42.
Wennerberg K, Ellerbroek SM, Liu RY, Karnoub AE, Burridge K, Der CJ.
J Biol Chem. 2002 Dec 6;277(49):47810-7. Epub 2002 Oct 9.
9.p35/cyclin-dependent kinase 5 phosphorylation of ras guanine nucleotide releasing factor 2 (RasGRF2) mediates Rac-dependent Extracellular Signal-regulated kinase 1/2 activity, altering RasGRF2 and microtubule-associated protein 1b distribution in neurons.
Kesavapany S, Amin N, Zheng YL, Nijhara R, Jaffe H, Sihag R, Gutkind JS, Takahashi S, Kulkarni A, Grant P, Pant HC.
J Neurosci. 2004 May 5;24(18):4421-31.
10.Rac1 is crucial for hair follicle integrity but is not essential for maintenance of the epidermis.
Chrostek A, Wu X, Quondamatteo F, Hu R, Sanecka A, Niemann C, Langbein L, Haase I, Brakebusch C.
Mol Cell Biol. 2006 Sep;26(18):6957-70.
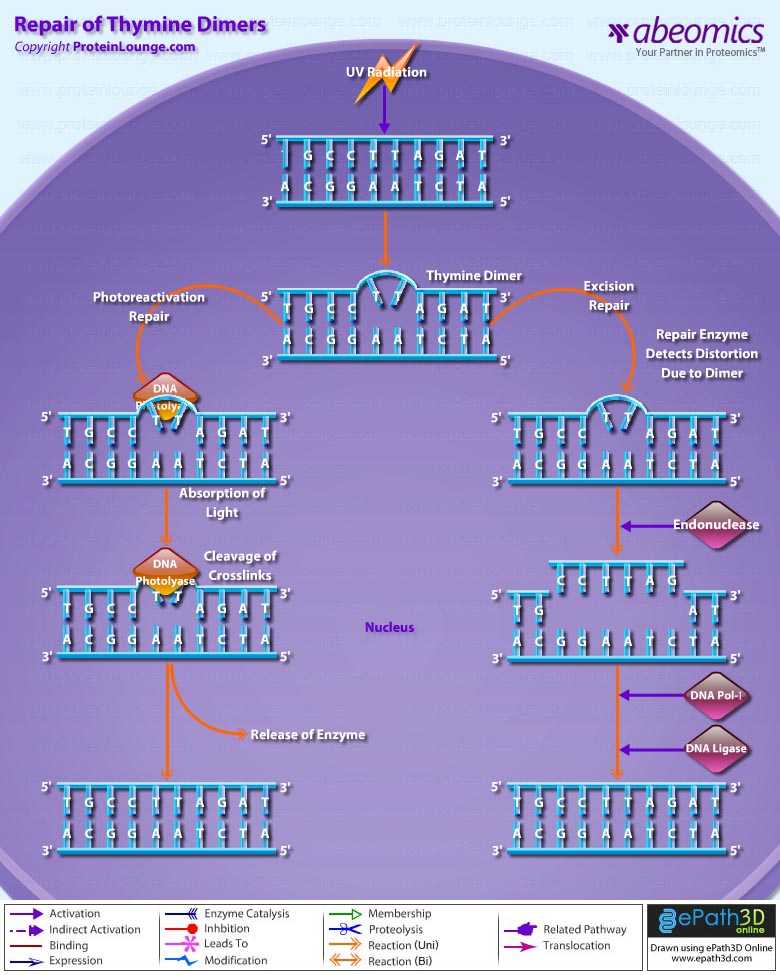
Irradiation of DNA by UV (Ultraviolet light) causes lesions, such as Cyclobutane-Pyrimidine Dimers or 6-4PPs (6-4 Pyrimidine Pyrimidone). The most common covalently linked adjoining pyrimidines are T-T (Thymine dimers), T-C (Thymine-Cytosine dimers) and C-C (Cytosine-Cytosine dimers). T-T dimers cause kinks in the DNA strand that prevent both replication and transcription of that part of the DNA. Because they block DNA replication (and therefore prevent cells from reproducing), T-T dimers and other forms of UV damage cannot be inherited, and thus do not constitute mutations. Such kinds of DNA damage are known as premutational lesions because they prevent both transcription and replication of the genes in which they are present and these lesions are fatal if they go uncorrected (Ref.2).
Several mechanisms are available for the removal or correction of T-T dimers from DNA depending upon the circumstances of the cell. Certain organisms posses a photoreactivating enzyme called Photolyase, which contain chromophores capable of capturing photons of blue light (of wavelength 350-450 nm). Photolyase first detect and bind to the damaged DNA site (a pyrimidine dimmer). Then it uses light energy absorbed from the visible range (lambda=370 nm) to oxidize the cyclobutane ring and convert the dimmer into monomers without disrupting the double strands. Finally, the enzyme dissociates from the DNA and the damage is repaired. Recognition of Cyclobutane-Pyrimidine Dimers by Photolyase is structure specific and the enzyme is not influenced by the nucleotide content surrounding the dimmer. Photoreactivation, however, is affected by the nucleotide content of the pyrimidine dimers. The Cyclobutane-Pyrimidine Dimers Photolyase is found in prokaryotes, lower and higher eukaryotes, but their existence in placental mammals is still unknown (Ref.1). Hence the repair of T-T dimers in humans takes place through excision repair mechanism. This repair mechanism does not require light and instead of just breaking the bonds of the T-T dimmer as was done by Photolyase, it excises the region of damaged nucleotides. A protein complex recognizes the distortion in the DNA caused by the T-T dimers and a pair of Endonucleases makes nicks in the DNA strand on either side of the T-T dimmer. Generally the nicks are 12 nucleotides apart and the DNA between the nicks is removed. DNA Polymerase-I fill in the gap left behind and DNA ligase seals the final nick in the DNA. In addition, other mechanisms such as mutagenic repair or dimmer bypass, recombinational repair, cell-cycle checkpoints, apoptosis and certain alternative repair pathways are also operative in various organisms for the removal or correction of T-T dimers (Ref.2).
The UV component of sunlight is responsible for the induction of skin tumors, most notably basal cell and squamous cell carcinomas and melanomas (Ref.3). The presence of UV induced dimers in the DNA is damaging and it may cause a mispairing as the strand is being copied or may stop replication altogether. Unless repaired, pyrimidine dimers may lead to blockage of transcription, mutations, cell death and cancer (Ref.4).
Chigancas V, Miyaji EN, Muotri AR, de Fatima Jacysyn J, Amarante-Mendes GP, Yasui A, Menck CF.
Cancer Res. 2000 May 1; 60(9): 2458-63.2.UV-induced DNA damage and repair: a review.
Sinha RP, Hader DP.
Photochem Photobiol Sci. 2002 Apr; 1(4): 225-36. Review.3.Photoreactivation of UV-induced cyclobutane pyrimidine dimmers in the MFA2 gene of Saccharomyces cerevisiae.
Morse NR, Meniel V, Waters R.
Nucleic Acids Res. 2002 Apr 15; 30(8): 1799-807.
4.Chromatin structure modulates DNA repair by photolyase in vivo.
Suter B, Livingstone-Zatchej M, Thoma F.
EMBO J. 1997 Apr 15; 16(8): 2150-60.
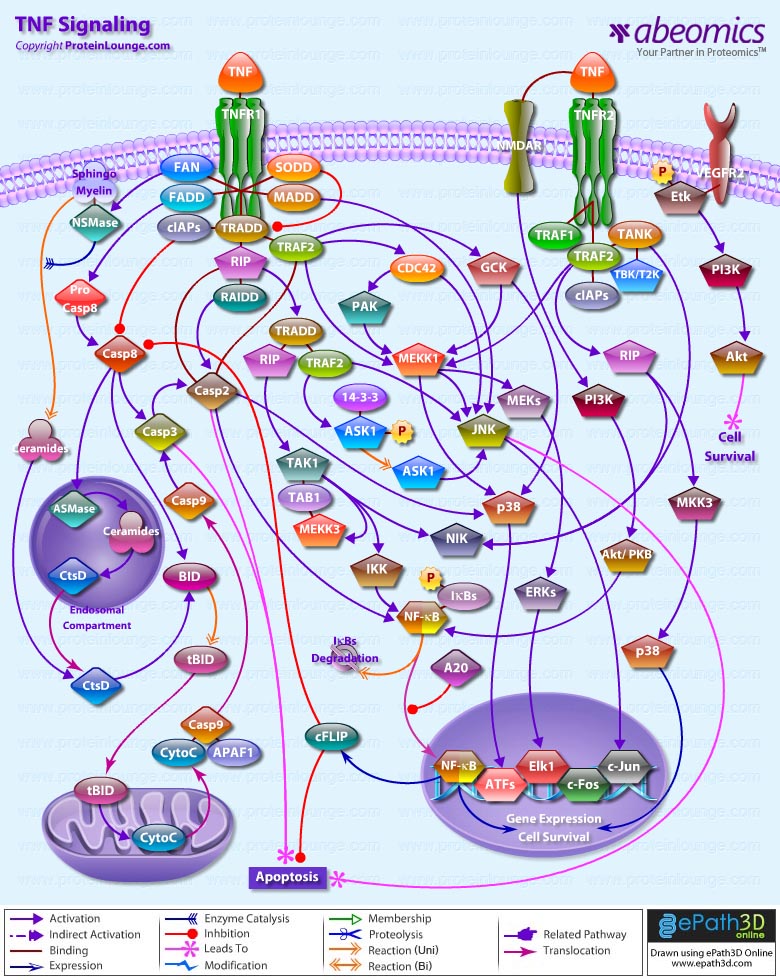
TNF (Tumor Necrosis Factor) is a multifunctional proinflammatory cytokine, with effects on lipid metabolism, coagulation, insulin resistance, and endothelial function. TNF has been considered as an anti-cancer agent since its discovery two decades ago. Members of the TNFR (TNF Receptor) super family can send both survival and death signals to cells. TNF family members play important roles in various physiological and pathological processes, including cell proliferation, differentiation, apoptosis, and modulation of immune responses and induction of inflammation. TNF acts through two receptors, TNFR1 (TNF Receptor-1) and TNFR2 (TNF Receptor-2). TNFR1 is expressed by all human tissues and is the major signaling receptor for TNF-Alpha. TNFR2 is mostly expressed in immune cells and mediates limited biological responses. TNFR2 binds both TNF-Alpha and TNF-Beta (Ref.1, 2, 3 & 4).
TNF-Alpha, a pro-inflammatory cytokine, is produced by many cell types, including macrophages, lymphocytes, fibroblasts and keratinocytes, in response to inflammation, infection, and other environmental stresses. TNF-Alpha induces a heterogeneous array of biological effects according to cell type. TNF-Alpha acts by binding to its receptors, TNFR1 (p55) and TNFR2 (p75), on the cell surface. Most cells express TNFR1, which is believed to be the major mediator of the cytotoxicity of TNF-Alpha (Ref.5 & 3). Binding of TNF-Alpha to its two receptors, TNFR1 and TNFR2, results in recruitment of signal transducers that activate at least three distinct effectors. Through complex signaling cascades and networks, these effectors lead to the activation of Caspases and two transcription factors, Activation Protein-1 and NF-KappaB (Nuclear Factor-KappaB). Initially, TRADD (TNFR-Associated Death Domain) protein, binds to TNFR1. Then, TRADD recruits FADD (Fas-Associated Death Domain), RAIDD (RIP-Associated ICH-1/CED-3-homologous protein with a Death Domain), MADD (MAPK Activating Death Domain) and RIP (Receptor-Interacting Protein). Binding of TRADD and FADD to TNFR1 leads to the recruitment, oligomerization, and activation of Caspase8. Activated Caspase8 subsequently initiates a proteolytic cascade that includes other Caspases (Caspases3, 6, 7) and ultimately induces apoptosis. Caspase8 also cleaves BID (BH3 Interacting Death Domain). tBID (Truncated BID) disrupts the outer mitochondrial membrane to cause release of the pro-apoptotic factors CytoC (Cytochrome-C). CytoC that is released from the intermembrane space binds to APAF1 (Apoptotic Protease Activating Factor-1), which recruits Caspase9 and in turn can proteolytically activate Caspase3. A novel 61-kDa protein kinase RIP2 is related to RIP that is a component of TNFR1 which mediates the recruitment of Caspase death proteases. Overexpression of RIP2 signaled both NF-KappaB activation and cell death (Ref. 5, 6, 3, 7, 8, & 9).
TRAF2 (TNF Receptor-Associated Factor-2) has been implicated in the activation of two distinct pathways that leads to the activation of Activation Protein-1 via the JNK (Jun NH2-terminal Kinase), MEKK (MEK Kinase), p38 and, together with RIP, NF-KappaB activation, via the NIK (NF-KappaB-Inducing Kinase). TNF-Alpha has been shown to activate MAPKs (Mitogen Activated Protein Kinases): ERK1 and ERK2 (Extracellular signal-Regulated Kinases). Overexpression of SODD (Silencer of Death Domains), a 60 kDa protein, associated with the DD of TNFRI suppresses TNF-induced cell death and NF-KappaB activation demonstrating its role as a negative regulatory protein for these signaling pathways.
TNFR2 is the receptor for TNF-Beta. TNF-Beta is produced by activated lymphocytes and can be cytotoxic to many tumor and other cells. In neutrophils, endothelial cells and osteoclasts TNF-Beta can lead to activation while in many other cell types it can lead to increased expression of MHC and adhesion molecules. TNF signaling has been implicated in many other diseases including: multiple sclerosis, Alzheimer’s disease, and TRAPS (TNF-Receptor-Associated Periodic Syndrome). A better understanding of TNF and its relatives should eventually result in the development of small molecules that can successfully inhibit and modulate the biological activity of these cytokines and thereby provide new avenues for therapeutic intervention (Ref. 10).
Raasch P, Schmitz U, Patenge N, Vera J, Kreikemeyer B, Wolkenhauer O.
BMC Bioinformatics. 2010 Sep 29;11:491.2.Azadirachtin interacts with the tumor necrosis factor (TNF) binding domain of its receptors and inhibits TNF-induced biological responses.
Thoh M, Kumar P, Nagarajaram HA, Manna SK.
J Biol Chem. 2010 Feb 19;285(8):5888-95. Epub 2009 Dec 14.3.TNF-mediated inflammatory disease.
Bradley JR.
J Pathol. 2008 Jan;214(2):149-60.
4.TNF signaling inhibition in the CNS: implications for normal brain function and neurodegenerative disease.
McCoy MK, Tansey MG.
J Neuroinflammation. 2008 Oct 17;5:45.
5.Cytokines in dermatology – a basic overview.
Coondoo A.
Indian J Dermatol. 2011 Jul;56(4):368-74.
6.Stakeholders’ perspectives on the regulation and integration of complementary and alternative medicine products in Lebanon: a qualitative study.
Alameddine M, Naja F, Abdel-Salam S, Maalouf S, Matta C.
BMC Complement Altern Med. 2011 Aug 28;11:71.
7.MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase.
Schievella AR, Chen JH, Graham JR, Lin LL.
J Biol Chem. 1997 May 2;272(18):12069-75.
8.Apoptosis signaling pathways and lymphocyte homeostasis.
Xu G, Shi Y.
Cell Res. 2007 Sep;17(9):759-71.
9.RIP2, a checkpoint in myogenic differentiation.
Munz B, Hildt E, Springer ML, Blau HM.
Mol Cell Biol. 2002 Aug;22(16):5879-86.
10.Tumor necrosis factor signaling.
Wajant H, Pfizenmaier K, Scheurich P.
Cell Death Differ. 2003 Jan;10(1):45-65.
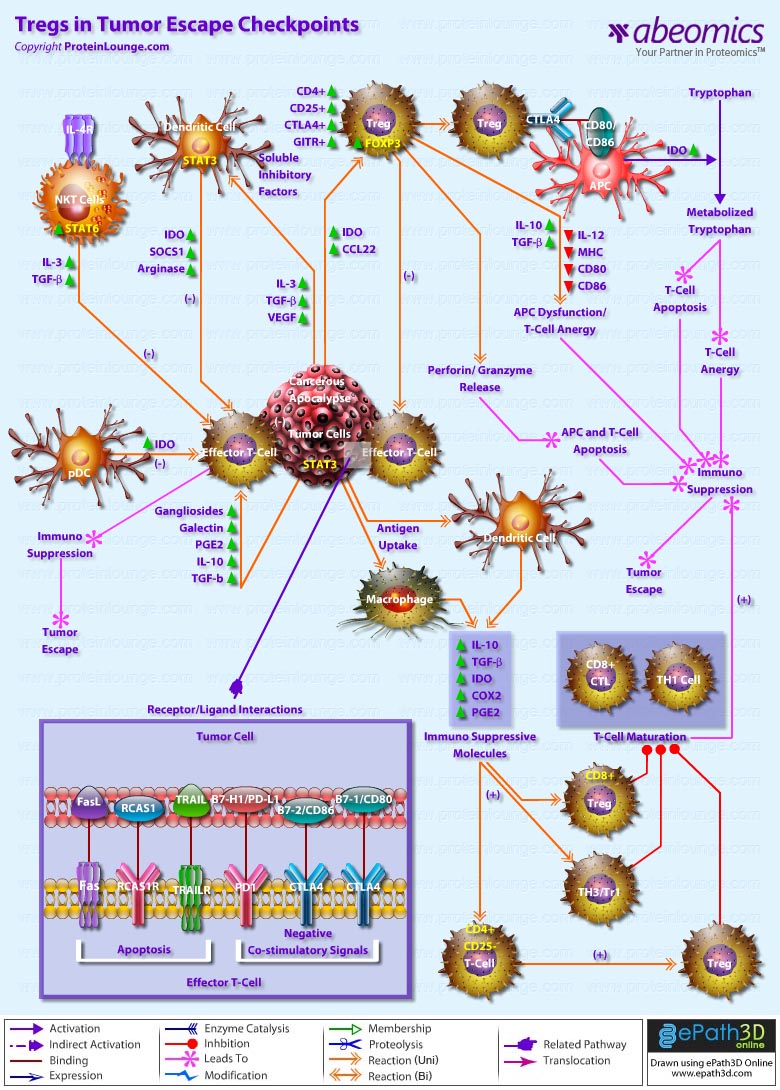
Specific T cell populations have suppressive/regulatory cells known as Regulatory T-cell or Tregs (Previously known as suppressor T-cell). Among them CD4+ regulatory T cells (Tregs) basically has two different subsets Tr1 and Th3 cells which are differentiated by their distinct suppressive mechanisms. The thymus-derived Tregs or natural Tregs (nTregs) express CD4 and high CD25 with FOXP3 (Forkhead Box-P3) to be a key regulatory gene (Ref.1). Apart from these, other markers of Treg include CTLA4 (Cytotoxic T-Lymphocyte-Associated protein-4), GITR (Glucocorticoid-Induced TNFR-Related protein) and LAG-3 (Lymphocyte Activation Gene-3). Treg cells are implicated in the development of autoimmunity, allergy and rejection of organ transplants, as well as the suppression of immune responses to cancer. Down regulation of the antigen presentation machinery has been considered as the most common strategy exploited by tumors cells to escape T cell control.
The key elements engaged in tumor escape include Treg cells, DCs (Dendritic cells) and NKT (Natural Killer Cells). CD4+ NKT cells suppress CTL (Cytotoxic T Lymphocytes)-effector functions by the secretion of IL-13 (Interleukin-13) and TGF-Beta (Transforming Growth Factor-Beta) which later activates IL-4R -STAT6 (Signal Transducer and Activator of Transcription-6) pathway (Ref.2&3). Tumor environmental factors, such as VEGF (Vascular Endothelial Growth Factor), IL-10 (Interleukin-10) and TGF-Beta suppress DCs differentiation and function, resulting in immature and/or partially differentiated DCs (Ref.4). These factors also induce STAT3 (Signal Transducer and Activator of Transcription-3) activation in DCs which in turn leads to impaired antigen-specific T cell levels of responses. These immature DCs dampens pre-existing antigen-specific effector T cell function in the presence of SOCS1 (Suppressors of cytokine signaling 1), Arginase and IDO (Indoleamine 2, 3-dioxygenase) (Ref.4, 5, 6&3). Again, pDCs (Plasmacytoid DCs) expresses the enzyme IDO that prevent the clonal expansion of T cells and promote T cell death. Tumor micro environmental CCL22 (Chemokine (C-C motif) Ligand-22) mediates trafficking of Treg cells into the tumor. In this process Treg cells express functional CCR4 (Chemokine (C-C motif) Receptor-4), the receptor for CCL22 , and migrate towards it in response to CCL22 in the tumor microenvironment, which is produced by tumor cells, in the presence of IDO and TGF-Beta . On activation, Treg cells proliferate and gain suppressive activity. Treg cells suppress both the innate and the adaptive immune response, including the priming and effector phase of both CD4 and CD8 T cells. Tumor-associated Macrophages and Dendritic cells also play a role in the expansion of Treg cells. Such expansion occurs through the proliferation of pre-existing Treg cells and the conversion of naive precursors into induced Treg cells in the presence of TGF-Beta and IDO (Ref.4, 5&7). Galectin-1 and Gangliosides along with TGF-Beta , PGE2 (Prostaglandin-E2) and IL-10 inhibits T cell effector functions by inducing T cell apoptosis (Ref.3). The suppressive molecules including VEGF, IL-10 , TGF-Beta , Arginase, IDO , PGE2, COX2 IL-12 and co signaling molecules such as CD80 or CD86 . These CTLs, when in sufficient abundance and directed against the appropriate tumor antigens, eradicate the tumor. Although the induced CTLs initially combat the tumor, they then fail as dysfunctional DC-induced Tregs ultimately inhibiting antitumor CTLs promoting tumor escape (Ref.8).
Treg cells within the tumor microenvironment are a crucial component of the tumor immunosuppressive network, the clinical depletion of Treg cells in patients with tumors can become a promising strategy for boosting TAA (Tumor-Associated Antigen)-specific immunity. Depletion of CD25+ cells using an antibody against CD25 results in improved tumor rejection. CTLA4 attenuates the potency of TAA-specific T-cell immunity. Administration of an antibody that blocks CTLA4 function inhibits the growth of moderately immunogenic tumors. Denileukin diftitox (Ontak) is a ligand toxin fusion, which depletes CD4+CD25+ TReg cells. Denileukin diftitox represents the first agent that is potentially useful for humans as blockade of Treg-cell-specific transport represents a novel means to augment tumor immunity (Ref.5).
Jonuleit H, Schmitt E.
J Immunol. 2003 Dec 15;171(12):6323-7. Review.2.NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway.
Terabe M, Matsui S, Noben-Trauth N, Chen H, Watson C, Donaldson DD, Carbone DP, Paul WE, Berzofsky JA.
Nat Immunol. 2000 Dec;1(6):515-20.3.Immunosuppressive strategies that are mediated by tumor cells.
Rabinovich GA, Gabrilovich D, Sotomayor EM.
Annu Rev Immunol. 2007;25:267-96. Review.
4.Regulatory T cells, tumour immunity and immunotherapy.
Zou W.
Nat Rev Immunol. 2006 Apr;6(4):295-307. Review.
5.Immunosuppressive networks in the tumour environment and their therapeutic relevance.
Zou W.
Nat Rev Cancer. 2005 Apr;5(4):263-74. Review.
6.Antigen-specific inhibition of effector T cell function in humans after injection of immature dendritic cells.
Dhodapkar MV, Steinman RM, Krasovsky J, Munz C, Bhardwaj N.
J Exp Med. 2001 Jan 15;193(2):233-8.
7.Regulatory-T-cell inhibition versus depletion: the right choice in cancer immunotherapy.
Colombo MP, Piconese S.
Nat Rev Cancer. 2007 Nov;7(11):880-7. Review.
8.Tregs and rethinking cancer immunotherapy.
Curiel TJ.
J Clin Invest. 2007 May;117(5):1167-74. Review.
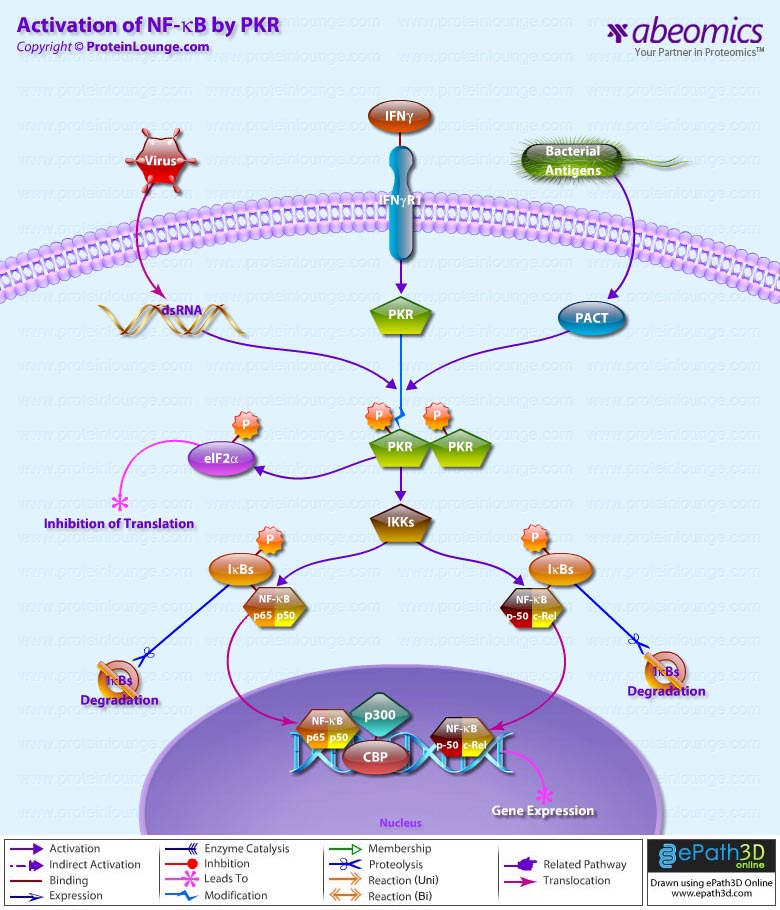
PKR (Protein Kinase-R) is a 68-kDa serine–threonine kinase that appears to play a primary role in mediating the antiviral activities of infected cells. PKR mediates apoptosis induced by many different stimuli, such as LPS (Lipopolysaccharides), TNF-Alpha (Tumour Necrosis Factor-Alpha), viral infection, or serum starvation. Viral infection leads to the increased expression and secretion of the cytokine IFN-Gamma (Interferon- Gamma) from host cells. IFN-gamma induces the dsRNA (Double-Stranded RNA)-dependent PKR. dsRNA, produced during viral replication, is an active component of a viral infection that stimulates antiviral responses in infected cells (Ref.1). The PKR protein is enzymatically inactive unless it is activated by binding to dsRNA. PKR activates as a kinase upon binding to dsRNA. Two DRBD (dsRNA Binding Domains) termed DRBDI and DRBDII, located in the NH2-terminal half of PKR, are necessary for its activation. The DRBD is a 70 amino acid motif that is conserved in a large family of dsRNA binding proteins and plays a key role in PKR function and regulation. Although their exact role in PKR activation is still not clear, the dsRNA binding domains might interact with dsRNA in a cooperative manner and contribute to optimal activation of PKR by facilitating PKR autophosphorylation and homodimerization. Binding to dsRNA induces PKR dimerization, autophosphorylation and activation. Once activated, PKR inhibits protein translation by phosphorylating the Alpha-subunit of the eIF2 (Eukaryotic Initiation Factor-2). Phosphorylation of eIF2-Alpha inhibits translation by sequestration of the guanine nucleotide exchange factor eIF2-Beta (Ref.2). In addition, PKR has been found to participate in transcriptional regulation of the E-selectin and immunoglobulin Kappa light chain genes. Besides, it also controls the activation of several transcription factors such as NF-KappaB (Nuclear Factor-KappaB), p53, or STATs (Signal Transducer and Activator of Transcription) (Ref.3)
Bacterial products or cellular stress can also activate PKR by an endogenous gene product called PACT (Protein Activator of Interferon-Induced Protein Kinase). The binding of PACT to PKR promotes conformational changes that allow PKR to activate the downstream signaling pathways leading to the activation of NF-KappaB (Nuclear Factor-KappaB). Rel/NF-KappaB transcription factors are a family of structurally related eukaryotic transcription factors that are involved in the control of a large number of normal cellular and organismal processes, such as immune and inflammatory responses, developmental processes, cellular growth, and apoptosis (Ref.4). In mammals, these proteins include p50 (NF-KappaB1), p52 (NF-KappaB2), p65 (RelA), RelB and c-Rel. These proteins share homology within a 300-amino-acid Rel homology domain, which mediates homo- and heterodimerization, DNA binding activity, and nuclear localization. A large number of stimuli including proinflammatory cytokines, antigen stimulation of T and B-Cells L, bacterial LPS, UV irradiation, ionizing radiation, viral infection, phorbol esters, reactive oxygen intermediates and bacterial or viral products can activate NF-KappaB and its target genes. NF-KappaB is found in the cytoplasm sequestered in a complex with I-KappaB (Inhibitor of Kappa Light Chain Gene Enhancer in B-Cells). Phosphorylation of I-KappaB triggers ubiquitin-dependent degradation, and subsequent release of NF-KappaB. NF-KappaB is then free to translocate to the nucleus to activate gene transcription (Ref.5).
PKR has been shown to activate NF-KappaB, either by directly phosphorylating I-KappaB, or indirectly by activating IKK (I-KappaB kinase). PKR physically interacts with IKK. IKK is a large multicomponent enzyme complex consisting minimally of three components: two closely related kinase subunits, IKK-Alpha and IKK-Beta, which have identical structural domains and interact as a heterodimer; and a third regulatory subunit termed IKK-Gamma (or NEMO). In the case of proinflammatory stimuli, including those that signal through PKR, activation of the IKK complex is dependent on IKK-Beta phosphorylation. PKR appears to associate with IKK through its catalytic domain, the dsRNA signal appears to be mediated through IKK-beta. Clearly, PKR is required for the efficient activation of NF-KappaB through the activation of IKK and degradation of I-KappaB. By phosphorylating I-KappaB, PKR functions within dsRNA- and IFN-signaling pathways to induce NF-KappaB-dependent transcription. NF-KappaB complexes induced by PKR are composed primarily of p50-p65 heterodimers and also of c-Rel-p50 heterodimers. The level of PKR activity in a cell is an important parameter contributing to cell fate decisions such as growth, differentiation and apoptosis. PKR has emerged as an important signaling mediator in a wide variety of cellular responses to extracellular stimuli. This kinase is required for the maintenance and propagation of signals necessary to sustain balanced host responses in proinflammatory and apoptotic situations (Ref.6).
References
1.PKR; a sentinel kinase for cellular stress.
Williams BR.
Oncogene. 1999 Nov 1;18(45):6112-20.
Williams BR.
Sci STKE. 2001 Jul 3;2001(89):RE2.
De Lucca FL, Serrano SV, Souza LR, Watanabe MA.
Eur J Pharmacol. 2002 Aug 16;450(1):85-9.
4.PACT, a protein activator of the interferon-induced protein kinase, PKR.
Patel RC, Sen GC.
EMBO J. 1998 Aug 3;17(15):4379-90.
5.The NF-kappa B and I kappa B proteins: new discoveries and insights.
Baldwin AS Jr.
Annu Rev Immunol. 1996;14:649-83.
6.PACT and PKR: turning on NF-kappa B in the absence of virus.
D’Acquisto F, Ghosh S.
Sci STKE. 2001 Jul 3;2001(89):RE1.
1.PKR; a sentinel kinase for cellular stress.
Williams BR.
Oncogene. 1999 Nov 1;18(45):6112-20.
Williams BR.
Sci STKE. 2001 Jul 3;2001(89):RE2.
De Lucca FL, Serrano SV, Souza LR, Watanabe MA.
Eur J Pharmacol. 2002 Aug 16;450(1):85-9.
4.PACT, a protein activator of the interferon-induced protein kinase, PKR.
Patel RC, Sen GC.
EMBO J. 1998 Aug 3;17(15):4379-90.
5.The NF-kappa B and I kappa B proteins: new discoveries and insights.
Baldwin AS Jr.
Annu Rev Immunol. 1996;14:649-83.
6.PACT and PKR: turning on NF-kappa B in the absence of virus.
D’Acquisto F, Ghosh S.
Sci STKE. 2001 Jul 3;2001(89):RE1.
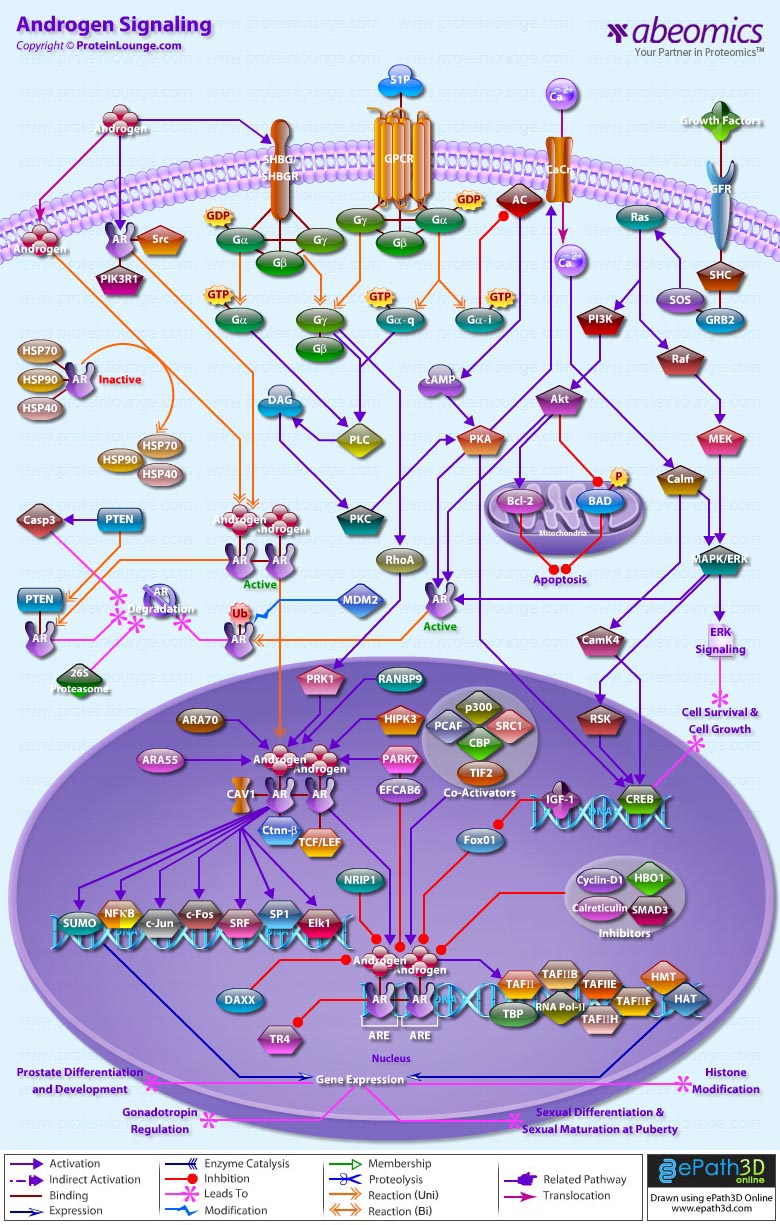
Androgens are recognized as genotropic inducers of a number of physiological functions mainly associated with the development of sexual characteristics. Androgens promote the growth and differentiation of prostate cells through ligand activation of the AR (Androgen Receptor) (Ref.1&2). The AR, upon activation by Androgens, mediates transcription of target genes that modulate growth and differentiation of prostate epithelial cells. AR signaling is crucial for the development and maintenance of male reproductive organs including the prostate gland (Ref.3).
The majority of serum Androgens is complexed with the plasma glycoprotein SHBG (Sex Hormone-Binding Globulin). The plasma protein SHBG binds to a receptor SHBGR (Sex Hormone-Binding Globulin Receptor) on cell membranes to form an SHBG- SHBGR complex. SHBGR transduces its signal through G proteins. Gs-protein coupled SHBG-R modulates AC (Adenylate-Cyclase) with synthesis of cAMP and activation of PKA (Protein Kinase A). PKA activation dephosphorylates AR resulting in its activation (Ref.4 & 5). PKA activation also significantly increases the phosphorylated form of its downstream effector, CREB (Camp Responsive Element-Binding Protein) in the presence of Androgen (Ref.6 & 7). In another signaling, S1P (Sphingosine-1-Phosphate) binds to and signals through several members of GPCRs (G Protein-Coupled Receptors) (Ref.8). The membrane-ARs transmit their signals via G-proteins. After agonist binding to its receptor, the G-alpha subunit releases GDP (Guanosine Diphosphate), binds GTP (Guanosine Triphosphate) and dissociates from the G-beta-gamma protein complex. G beta-gamma subunits cannot be dissociated under non-denaturing conditions, and regulate activity of PLC (Phospholipase C). Activation of PLC causes increased IP3 (Inositol Trisphosphate) and DAG (Diacyl Glycerol) through PIP2 (Phosphatidylinositol-4, 5-Biphosphate) and DAG activates PKC (Protein kinase C) (Ref.4). PKC, on activation, activates PKA which in turn activates Ca2+ channels. The major routes to regulate the intracellular Ca2+ are plasma membrane integrated Ca2+-channels by L-/T-type Ca2+-channels and sarcoplasmic reticulum by IP3-receptors (Ref.4). Androgens stimulate an elevation in intracellular Ca2+ through a G protein-coupled receptor. The elevation of intracellular Ca2+ is detected by specific Ca2+ sensor molecules, including PKC and CaM (Calmodulin), to induce signal transduction cascades and modulation of transcription factor activity. Simultaneously IP3 activates specific receptors in the sarcoplasmic reticulum to release Ca2+ into the cytosol (Ref.5 & 9). Ca2+ functions as a ubiquitous messenger and modulation of intracellular Ca2+ levels regulates a wide range of cellular processes, including proliferation, apoptosis, motility, and gene expression. The Androgen–AR complex interacts with the SH3 domain of Src (Steroid Receptor Coactivator) kinase and leads to a rapid activation of Shc (SHC (Src Homology-2 Domain Containing) Transforming Protein) which rapidly induce the formation of SHC/GRB2 (Growth Factor Receptor-Binding Protein 2)/SOS (Son Of Sevenless) complexes, leading to the stimulation of the MAPK (Mitogen-Activated Protein Kinase) pathway, involving growth factors and growth factor receptors. In cooperation with GRB2, the guanine-nucleotide exchange factor SOS potentiates Ras, by catalyzing the replacement of GDP for GTP. Ras activates Raf-1 which further stimulates MAPK mediating MEK (Mitogen-Activated Protein/Extracellular Signal-Regulated Kinase). In another related signaling, Ras recruits PI3K (Phosphatidylinositol 3-Kinase) phosphorylates and potentiates Akt (v-Akt Murine Thymoma Viral Oncogene Homolog-1) (Ref.4&5)). MAPK directly modifies ligand-dependent activation of AR modulating AREs (Androgen-Receptor Element)-inducible gene transcription downstream of the AR. CREB is a downstream target of active MAPK through the mediation of the RSK (RSK kinase) and p300/CBP (CREB-Binding Protein) bridge the interaction between AR and CREB (Ref.6&10). Activation of the PI3K–Akt pathway promotes AR ubiquitylation and leading to AR degradation via a proteasome-dependent pathway. MDM2 (Mouse Double Minute-2) and Akt form a complex with AR and induce AR ubiquitylation and degradation in a phosphorylation-dependent manner. AR phosphorylation by Akt promotes AR ubiquitylation and subsequent degradation by the 26S proteasome (Ref.11). Phosphorylation of Bcl2 by Raf-1-ERK dependent pathway enhances its anti-apoptotic function. Moreover, Bcl2(B-cell Lymphoma Protein-2) acts dimerizing with and inhibiting pro-apoptotic protein counterparts such as BAD (Bcl2-Antagonist of Cell Death) and the phosphorylation of Bcl2 likely favors the dimerization with BAD. The phosphorylation of BAD is followed by its inactivation (Ref.12). A triple complex between AR, the regulatory subunit p85 of PI3K (PIK3R1) and Src tyrosine kinase is formed in the presence of Androgen, which is negatively regulated by PTEN (Phosphatase and Tensin Homologue) as it function as an AR corepressor. The direct interaction between the AR and PTEN inhibits AR nuclear translocation and promotes AR protein degradation (Ref.7). PTEN also cleaves Caspases 8, 9 and 3 (Ref.13). G12 regulates low molecular weight GTPase RhoA, but when signaling, G-proteins function as dimers because the signal is communicated either by the G-alpha subunit or the G-beta-gamma complex (Ref.4). A signal transduction pathway links the RhoA effector PRK1 (Protein Kinase C-Related Kinase-1) to the transcriptional activation of the AR (Ref.14). Further, in the AR activation process, AR LBD (Ligand Binding Domain) interacts transiently with HSP40 (Heat Shock Proteins-40), HSP70, HSP90. HSP70 and HSP40 also reassociate with the AR in the presence of ligand and facilitate transport of the receptor into the nucleus and this process is accompanied by coregulators of AR, such as ARA55 (Androgen Receptor Coactivator-55) and ARA70 (Ref.7). In the Wnt signaling, Wnt binds to its receptors Frizzled and LRP5/6, activating the downstream component Dishevelled, which in turn inhibits GSK-3â (Glycogen Synthetase Kinase-3Beta), Axin and APC (Adenomatous Polyposis Coli) in the â-Catenin destruction complex. Once stabilized, â-Catenin binds to LEF (Lymphoid Enhancer Factor)/TCF (T-Cell Factor) transcription factors in the nucleus, interacting with the AR and potentiating the AR transcriptional activity in an Androgen-dependent fashion. At the same time, GSK-3â interacts with the AR and suppresses its ability to activate transcription whereas on the other side Akt phosphorylates and inactivates GSK-3â (Ref.7&15)). In the nucleus, DJ-1 interacts directly with the AR whereas DJBP (DJ-1-Binding Protein) binds the DBD of the AR in an Androgen-dependent manner and co-localizes with DJ-1. DJBP represses Androgen-dependent AR transactivation activity by recruiting a HDAC complex. DJ-1 partially restores the activity of the AR by abrogating the DJBP-HDAC complex (Ref.7&16)). Among the other negative regulators of AR, includes Calreticulin, HBO1 (Human Origin Recognition Complex Interacting Protein), Daxx (Death-Domain Associated Protein), Smad3 (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-3)), DAX-1 (DSS-AHC Critical Region on The X Chromosome Protein-1) and cyclin D1 that repress ligand-mediated transcriptional activation of AR. Again, FOXO1 (Forkhead Box O1), as a downstream molecule becomes phosphorylated and inactivated by PI3K/AKT kinase in response to growth factors, and subsequently suppresses ligand mediated AR transactivation. NRIP1 (Nuclear Receptor Interacting Protein 1) also act as a strong AR repressor. NRIP1 reverse TIF-2 (Transcriptional Intermediary Factor-2) dependent overactivation of AR. SUMO-1(Small Ubiquitin-Related Modifier) decreases, whereas SUMO-2 and -3 enhance AR transcriptional activity. AR is covalently modified by SUMO-1 (sumoylated) in an Androgen-enhanced fashion. Whereas, with TR4 (Testicular Orphan Receptor-4), AR interact with it and function as a repressor to down-regulate the TR4 target genes by preventing the TR4 binding to its target DNA (Ref.2, 7, 17&18). The formation of a productive AR transcriptional complex requires the functional and structural interaction of the AR with its coregulators as, CBP, p300, P/CAF (P300/CBP-Associated Factor), SRC-1(Steroid Receptor Co-Activator-1), TIF-2 (Transcriptional Intermediary Factor-2), which contain inherent HAT (Histone Acetylation) activity. Moreover, RanBP9 (Ran-Binding Protein) and HIPK3 (Homeodomain-Interacting Protein Kinase 3) are identified as AR-binding protein. AR interacts with other transcription factors, including NF-kB(Nuclear Factor-kB ), SRF (Serum Response Factor), sp1 (Specificity Protein 1), c-Jun and c-Fos (Cellular Oncogene Fos). An increased activity of another transcription factor Elk-1 also activates c-fos expression and elicits gene transcription independently of AR binding to DNA response elements (Ref. 7, 18&19). Enhanced transcription by the AR depends on the recruitment of RNA polymerase II to promoters of its target genes. This is achieved by the assembly of general transcription factors that make up the PIC (Preinitiation Complex). Formation of the PIC is accomplished by binding of TFIID, which is composed of TBP (TATA-binding protein) and TBP associated factors, in the proximity of the transcriptional start site. TFIIB then binds TBP and recruits RNA polymerase II and TFIIF, which ensures specific interaction of RNA polymerase II at the promoter. TFIIE and TFIIH are recruited to RNA polymerase II to facilitate strand separation, which allows transcription initiation. AR as well binds to HMT (Histone Methyltransferase) in this process (Ref. 7).
AR point mutations changes expression of AR co-regulatory proteins leading to prostate cancers and more frequently tumors. Significant lower levels of Androgen causes CAD (coronary artery disease), thus implying that low circulating Androgen levels are correlated with progression of atherosclerosis. Indeed, numerous risk factors for CAD are associated with Hypotestosteronaemia. Chronic heart failure due to idiopathic dilated cardiomyopathy is also associated with a significant decrease in Androgen level. In neurons, Androgens are also able to rapidly modulate Neuronal synaptic plasticity (Ref.4&20).
References
1.Androgens are powerful non-genomic inducers of calcium sensitization in visceral smooth muscle.
González-Montelongo MC, Marín R, Gómez T, Díaz M.
Steroids. 2009 Oct 1.
2.Androgen receptor and growth factor signaling cross-talk in prostate cancer cells.
Zhu ML, Kyprianou N.
Endocr Relat Cancer. 2008 Dec;15(4):841-9.
3.Targeting the androgen receptor pathway in prostate cancer.
Chen Y, Sawyers CL, Scher HI.
Curr Opin Pharmacol. 2008 Aug;8(4):440-8.
Michels G, Hoppe UC.
Front Neuroendocrinol. 2008 May;29(2):182-98.
5.The roles of androgen receptors and androgen-binding proteins in nongenomic androgen actions.
Heinlein CA, Chang C.
Mol Endocrinol. 2002 Oct;16(10):2181-7. Review.
Kim J, Jia L, Stallcup MR, Coetzee GA.
J Mol Endocrinol. 2005 Feb;34(1):107-18.
Heemers HV, Tindall DJ.
Endocr Rev. 2007 Dec;28(7):778-808.
8.Homodimerization and heterodimerization of S1P/EDG sphingosine-1-phosphate receptors.
Van Brocklyn JR, Behbahani B, Lee NH.
Biochim Biophys Acta. 2002 May 23;1582(1-3):89-93. Review.
9.Non-genomic actions of androgens.
Foradori CD, Weiser MJ, Handa RJ.
Front Neuroendocrinol. 2008 May;29(2):169-81.
Unni E, Sun S, Nan B, McPhaul MJ, Cheskis B, Mancini MA, Marcelli M.
Cancer Res. 2004 Oct 1;64(19):7156-68.
Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C.
EMBO J. 2002 Aug 1;21(15):4037-48.
Caraglia M, Marra M, Leonetti C, Meo G, D’Alessandro AM, Baldi A, Santini D, Tonini G, Bertieri R, Zupi G, Budillon A, Abbruzzese A.
J Cell Physiol. 2007 May;211(2):533-43.
13.Human sperm express a functional androgen receptor: effects on PI3K/AKT pathway.
Aquila S, Middea E, Catalano S, Marsico S, Lanzino M, Casaburi I, Barone I, Bruno R, Zupo S, Andò S.
Hum Reprod. 2007 Oct;22(10):2594-605.
Metzger E, Müller JM, Ferrari S, Buettner R, Schüle R.
EMBO J. 2003 Jan 15;22(2):270-80.
Terry S, Yang X, Chen MW, Vacherot F, Buttyan R.
J Cell Biochem. 2006 Oct 1;99(2):402-10.
Niki T, Takahashi-Niki K, Taira T, Iguchi-Ariga SM, Ariga H.
Mol Cancer Res. 2003 Feb;1(4):247-61.
17.Receptor-interacting protein 140 is a repressor of the androgen receptor activity.
Carascossa S, Gobinet J, Georget V, Lucas A, Badia E, Castet A, White R, Nicolas JC, Cavaillès V, Jalaguier S.
Mol Endocrinol. 2006 Jul;20(7):1506-18.
18.Negative modulation of androgen receptor transcriptional activity by Daxx.
Lin DY, Fang HI, Ma AH, Huang YS, Pu YS, Jenster G, Kung HJ, Shih HM.
Mol Cell Biol. 2004 Dec;24(24):10529-41.
Yemelyanov A, Czwornog J, Gera L, Joshi S, Chatterton RT Jr, Budunova I.
Cancer Res. 2008 Jun 15;68(12):4763-73.
20.Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence.
Taplin ME, Balk SP.
J Cell Biochem. 2004 Feb 15;91(3):483-90.
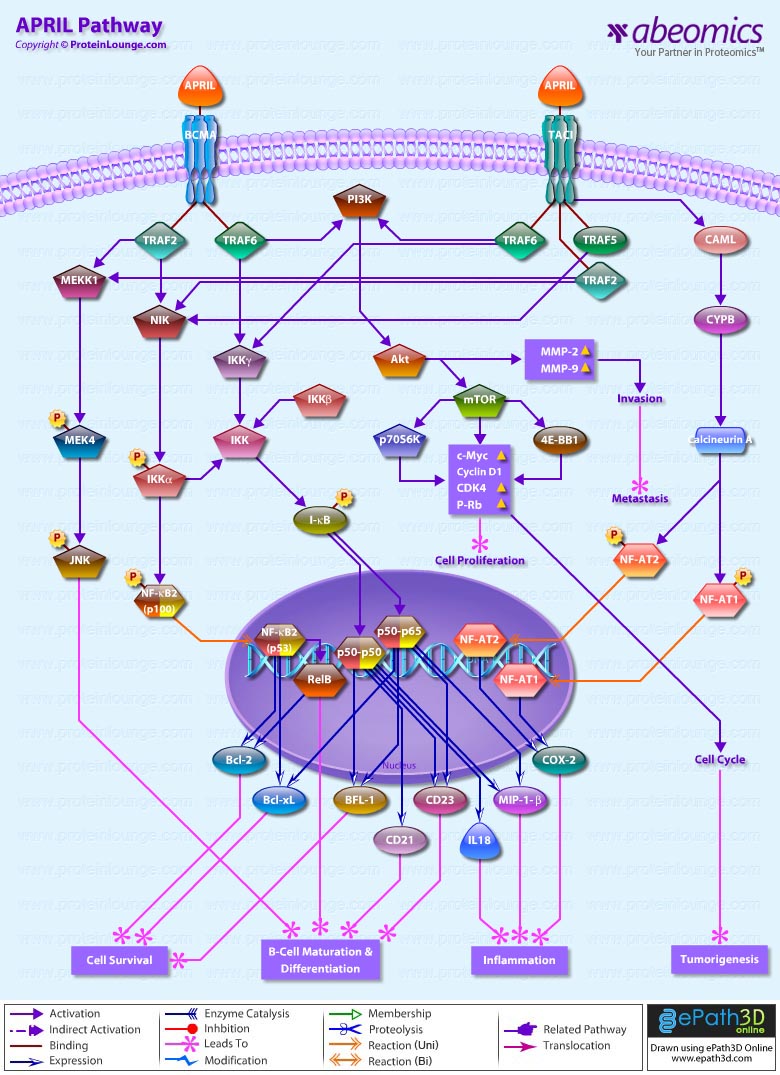
The TNF (Tumor Necrosis Factor) and TNF receptor superfamilies (TNFSF and TNFRSF) consist of approximately 50 membrane and soluble proteins that can modulate cellular function. Most of these molecules are expressed by or can target cells of the immune system, and they have a wide range of actions including promoting cellular differentiation, survival, and production of inflammatory cytokines and chemokines. Members of the TNFRSF (Tumor Necrosis Factor Receptor superfamily ) participate prominently in B-cell maturation and function. In particular, BAFFR (B-cell Activating Factor) belonging to the TNF family receptor, BCMA (B-Cell Maturation Antigen), and transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI) play critical roles in promoting B-cell survival at distinct stages of development by engaging a proliferation-inducing ligand APRIL (A Proliferation-Inducing Ligand) and/or BAFF (Ref.1).
APRIL, also known as TALL2 (TNF- and APOL-related Leukocyte expressed Ligand-2), has no essential function in normal B cell development and plays only a minor role in B cell homeostasis. In immune responses APRIL acts as a costimulator for B and T cell proliferation and supports class switch. In general, cytokines of the TNF superfamily are functional both in their membrane-bound and in soluble form when shed by enzymatic cleavage from the surface through a furin-like convertase. APRIL is secreted following intracellular processing in the Golgi apparatus. APRIL binds to two separate receptors, the TACI (Transmembrane Activator and Calcium modulator ligand Interactor), and the BCMA (B Cell Maturation Antigen). Both receptors are expressed on B cells, although their expression level changes with B cell maturation (Ref. 2). These receptors are functional as their ligation triggers activation of NF-KappaB, MAPK/JNK, and Akt signaling. APRIL may also induce apoptosis through its interaction with other TNF receptor family proteins such as TNFRSF6/FAS and TNFRSF14/HVEM.
Signaling through TACI in mature B cells or plasmablasts are only achieved by higher-order APRIL oligomers, all of which were also potent activators of a multimerization-dependent reporter signaling pathway. TACI intracellular domain interacts with TNFR-associated factor TRAF2, 5, and 6. TRAFs are trimeric intermediates in the signaling pathway of numerous TNF receptor family members. By bringing together the receptors, a trimeric ligand may allow recruitment to the complex of just one intracellular TRAF, which may not be enough to induce signaling, as oligomerization of trimeric TRAF-2 and TRAF-6 is required to activate downstream signaling events (Ref. 3). Cytoplasmic domain of TACI encompasses a conserved motif that bound MyD88, an adaptor that activates transcription factor NF-?B signaling pathways via a Toll–interleukin 1 (IL-1) receptor (TIR) domain. TACI lacks a TIR domain, yet triggered CSR via the DNA-editing enzyme AID by activating NF-?B through a Toll-like receptor (TLR)-like MyD88-IRAK1-IRAK4-TRAF6-TAK1 pathway (Ref. 4 and 5). BCMA binds to APRIL with high affinity. BCMA enhances B cell antigen presentation by recruiting TRAF proteins and activating NF-KappB. BCMA might regulate post-CSR events, including the differentiation and survival of mucosal IgA-secreting plasma cells (Ref. 6).
References
1.Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease.
Rickert RC, Jellusova J, Miletic AV.
Immunol Rev. 2011 Nov;244(1):115-33. doi: 10.1111/j.1600-065X.2011.01067.x.
2.In vitro and in vivo activation induces BAFF and APRIL expression in B cells.
Chu VT, Enghard P, Riemekasten G, Berek C.
J Immunol. 2007 Nov 1;179(9):5947-57.
Bossen C, Cachero TG, Tardivel A, Ingold K, Willen L, Dobles M, Scott ML, Maquelin A, Belnoue E, Siegrist CA, Chevrier S, Acha-Orbea H, Leung H, Mackay F, Tschopp J, Schneider P.
Blood. 2008 Feb 1;111(3):1004-12. Epub 2007 Oct 17.
He B, Santamaria R, Xu W, Cols M, Chen K, Puga I, Shan M, Xiong H, Bussel JB, Chiu A, Puel A, Reichenbach J, Marodi L, Döffinger R, Vasconcelos J, Issekutz A, Krause J, Davies G, Li X, Grimbacher B, Plebani A, Meffre E, Picard C, Cunningham-Rundles C, Casanova JL, Cerutti A.
Nat Immunol. 2010 Sep;11(9):836-45. Epub 2010 Aug 1.
Xia XZ, Treanor J, Senaldi G, Khare SD, Boone T, Kelley M, Theill LE, Colombero A, Solovyev I, Lee F, McCabe S, Elliott R, Miner K, Hawkins N, Guo J, Stolina M, Yu G, Wang J, Delaney J, Meng SY, Boyle WJ, Hsu H.
J Exp Med. 2000 Jul 3;192(1):137-43.
6.Innate control of B cell responses.
Cerutti A, Puga I, Cols M.
Trends Immunol. 2011 May;32(5):202-11. Epub 2011 Mar 16. Review.
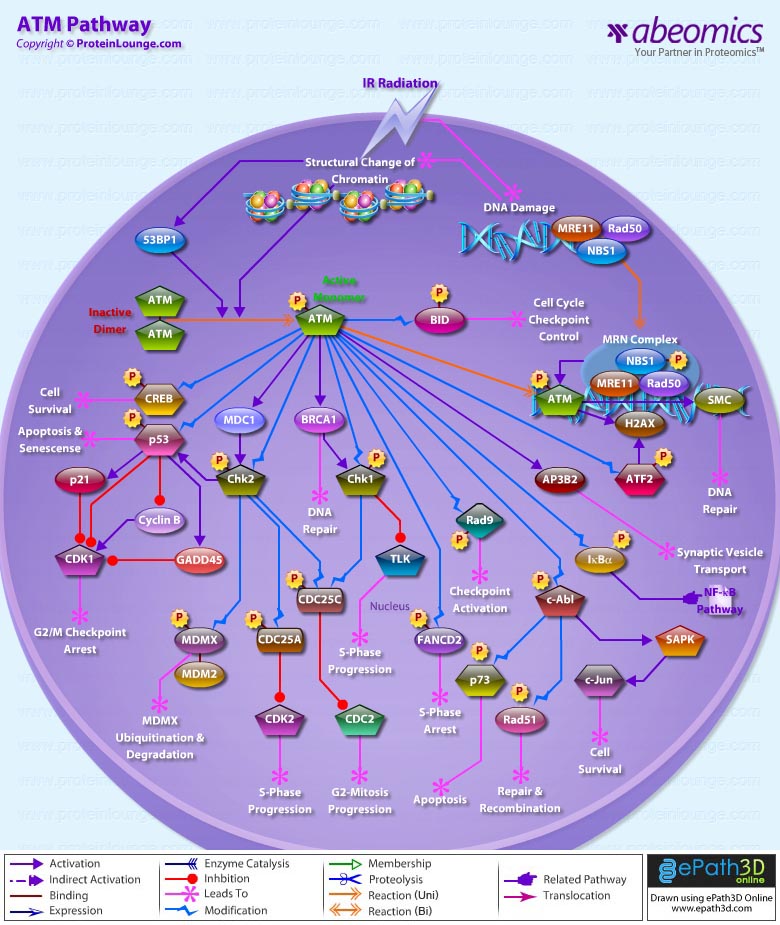
ATM (Ataxia Telangiectasia Mutated Protein) belongs to a family of Kinases that have sequence homology to PI3K (Phosphoinositide 3-Kinase). ATM is a key regulator of multiple signaling cascades which respond to DNA strand breaks induced by damaging agents IR (Ionizing Radiation), radiometric agents or by normal processes. These responses involve the activation of cell cycle Chk factors (Checkpoints factors), DNA repair and Apoptosis. In addition, ATM appears to function as a ‘caretaker’, suppressing tumorigenesis in specific T cell lineages. Its downstream targets include Chk1 (Cell Cycle Checkpoint Kinase-1), Chk2 (Cell Cycle Checkpoint Kinase-2), tumor suppressors like p53 and BRCA (Breast Cancer), DNA repair factors like Rad50, Rad51, GADD45 (Growth Arrest and DNA-Damage-inducible), and other signaling molecules like c-Abl and NF-KappaB (Nuclear Factor-Kappa B) (Ref.1). In non-irradiated cells ATM exists as a dimer, is not phosphorylated and is present throughout the nucleus. After irradiation, which leads to formation of DNA DSBs (DNA Double Strand Breaks), ATM becomes a monomer, it is phosphorylated on Ser1981 and some pool of it is present at sites of DNA DSBs. ATM monitors the presence of DNA DSBs indirectly, through DNA DSB-induced changes in chromatin structure. ATM is activated by the MRE11 (Meiotic Recombination-11)–Rad50–NBS1 (Nijmegen Breakage Syndrome-1) complex or 53BP1. The former is thought to be recruited at the DSB, whereas the latter is recruited at chromatin regions flanking the DNA DSB and extending up to a few megabases from the DSB. ATM phosphorylates NBS1 on several residues in response to DNA damage and a functional MRE11/Rad50/NBS1 complex is required for Chk2 activation. This links the MRE11/Rad50/NBS1 complex to DNA damage recognition. ATM activated at sites of DNA DSBs may also phosphorylate and activate ATM in the nucleoplasm (Ref.1 & 2).
Once ATM is activated it phosphorylates multiple substrates. Two of these, Chk2 and p53, mediate many of the cell cycle effects of ATM, while two others, SMC1 (Structural Maintainance of Chromosomes-1) and histone H2AX (H2A Histone family, member X), are important for cell survival after irradiation. Chk2 is a protein Kinase, that once activated amplifies the DNA damage signal of ATM. Two of the key substrates of Chk2 are CDC25A (Cell Division Cycle-25A) and CDC25C (Cell Division Cycle-25C). CDC25A and CDC25C are protein phosphatases; CDC25A activates CDK2 and promotes progression through S phase, while CDC25C activates CDC2 and promotes progression from G2 into mitosis. When CDC25A and CDC25C become phosphorylated by Chk2, their function is inhibited and cells delay progression through S phase or arrest in G2. Chk2 contributes to p53 regulation in part through phosphorylation of MDMX. The mechanism by which phosphorylation enhances MDMX–MDM2 (Mouse Double Minute-2) binding and MDMX ubiquitination by MDM2 remains to be further investigated. MDC1 (Mediator of DNA damage Checkpoint-1) localizes to sites of DNA breaks and associates with Chk2 after DNA damage. This association is mediated by the MDC1 FHA domain and the phosphorylated Thr 68 of Chk2. Cell cycle arrest in G1 is mediated by p53, which is a substrate of both Chk2 and ATM. p53 is a transcription factor that induces expression of p21 (Cyclin Dependent Kinase Inhibitor-p21)/Waf1, GADD45, and MDM2. p21 is a CDK(Cyclin-Dependent Kinase) inhibitor. p53 also inhibits G2-M transition by repressing the transcription of CDC2 (Cell Division Cycle-2, G1 to S and G2 to M) and Cyclin-B. p53 can also induce expression of genes that induce apoptosis; and in certain tissues, induction of p53 leads to apoptosis, rather than cell cycle arrest. Unlike, Chk2 and p53, Phosphorylated SMC1 and Histone H2AX are found exclusively at sites of DNA DSBs. SMC1 phosphorylation is critical for cells to survive after irradiation. Similar to SMC1 phosphorylation, histone H2AX phosphorylation also appears to be important for DNA repair. The mechanisms by which SMC1 and histone H2AX phosphorylation facilitate DNA repair are not known, but recent evidence suggests that phosphorylated histone H2AX recruits chromatin remodeling complexes to sites of DNA DSBs (Ref.3, 4 & 5).
ATM also interacts with the protein product of the c-Abl proto-oncogene. This non-receptor tyrosine Kinase is involved in several stress responses including activation of the SAPK (Stress-Activated Protein Kinase), Rad51 and p73. SAPK activation by c-Abl leads to activation of c-Jun and thus plays an important role in cell survival. c-Abl also phosphorylates Rad51, which is further involved in DNA repair and recombination processes. Activated c-Abl may also promote apoptosis via up-regulation of p73, a proapoptotic protein and a p53 homolog. ATM also interacts with AP3B2 (Adaptor-related Protein complex-3, Beta-2 subunit), a neuronal homolog of Beta-Adaptin thought to be involved in synaptic vesicle transport in neuronal cells. ATM phosphorylates ATF2 (Activating Transcription Factor-2) on Serine 490 and 498 following IR (Ionizing Radiation). Phosphorylation of ATF2 by ATM results in its rapid colocalization with Gamma-H2AX and MRN components into IRIF (IR-Induced Foci). ATM also phosphorylates IKappaB-Alpha and thus plays an important role in NF-KappaB activation. ATM also phosphorylates and activates Chk1, which phosphorylates TLK1 (Tousled Like Kinase-1). TLK activity is rapidly suppressed by DNA damage and by inhibitors of replication. Chk1 also phosphorylate a conserved site (Ser-216) on protein phosphatase CDC25C, which results in it being inactivated and bound by the 14-3-3 protein. The inactive CDC25C is then incapable of removing an inhibitory phosphate group on Tyr-15 of CDC2, preventing entry into mitosis. ATM also phosphorylates FANCD2 (Fanconi Anemia Complementation Group-D2) protein, which then leads to S phase arrest (Ref.5, 6 & 7).
BRCA1 is also activated by the protein kinase ATM that initiates cell cycle changes after DNA damage. BRCA1 physically interacts with Chk1 and stimulates its activity after DNA damage. Hyperphosphorylation of Rad9 induced by IR is also dependent on ATM. Ser(272) of Rad9 is phosphorylated directly by ATM. ATM also phosphorylates CREB (cAMP Response Element-Binding Protein-1) in vitro and in vivo in response to IR and H2O2 on a stress-inducible domain. ATM-mediated phosphorylation of CREB in response to DNA damage modulates CREB-dependent gene expression and dysregulation of the ATM-CREB pathway may contribute to neurodegeneration. ATM also phosphorylates BID (BH3 Interacting Domain Death Agonist). This phosphorylation is required for a downstream function in cell cycle checkpoint control. ATM also seems to have a role in telomere maintenance and replication. The disease AT (Ataxia-Telangiectasia), caused by mutations in the ATM gene, is an autosomal recessive disorder characterized by Progressive Cerebellar Ataxia, Cancer Predisposition, Oculocutaneous Telangiectasia and variable immune deficiencies. Inhibiting ATM might sensitize repair proficient cancer cells to radiation and chemotherapy. It is possible that targeting the ATM pathway might even introduce selectivity to current cancer therapy. However, the real test of this hypothesis will have to await the development of more potent and specific inhibitors for ATM (Ref.8, 9 & 10).
References
1.Exclusion/confirmation of Ataxia-telangiectasia via cell-cycle testing.
Heinrich T, Prowald C, Friedl R, Gottwald B, Kalb R, Neveling K, Herterich S, Hoehn H, Schindler D.
Eur J Pediatr. 2006 Jan 13;:1-8 [Epub ahead of print]
Zgheib O, Huyen Y, DiTullio RA Jr, Snyder A, Venere M, Stavridi ES, Halazonetis TD.
Radiother Oncol. 2005 Aug;76(2):119-22.
3.DNA damage regulates CHK2 association with chromatin.
Li J, Stern DF.
J Biol Chem. 2005 Sep 8; [Epub ahead of print]
4.ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation.
Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG.
Cell Cycle. 2005 Sep;4(9):1166-70. Epub 2005 Sep 29.
5.Defective ATM-p53-mediated apoptotic pathway in multiple sclerosis.
Deng X, Ljunggren-Rose A, Maas K, Sriram S.
Ann Neurol. 2005 Oct;58(4):577-84.
6.ATM-dependent phosphorylation of ATF2 is required for the DNA damage response.
Bhoumik A, Takahashi S, Breitweiser W, Shiloh Y, Jones N, Ronai Z.
Mol Cell. 2005 May 27;18(5):577-87.
Krause DR, Jonnalagadda JC, Gatei MH, Sillje HH, Zhou BB, Nigg EA, Khanna K.
Oncogene. 2003 Sep 4;22(38):5927-37.
8.Direct regulation of CREB transcriptional activity by ATM in response to genotoxic stress.
Shi Y, Venkataraman SL, Dodson GE, Mabb AM, LeBlanc S, Tibbetts RS.
Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):5898-903. Epub 2004 Apr 8.
9.Proapoptotic BID is an ATM effector in the DNA-damage response.
Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, Haimovich G, Lerenthal Y, Marcellus RC, Gross A.
Cell. 2005 Aug 26;122(4):593-603.
10.ATM and ataxia telangiectasia.
McKinnon PJ.
EMBO Rep. 2004 Aug;5(8):772-6. Review.
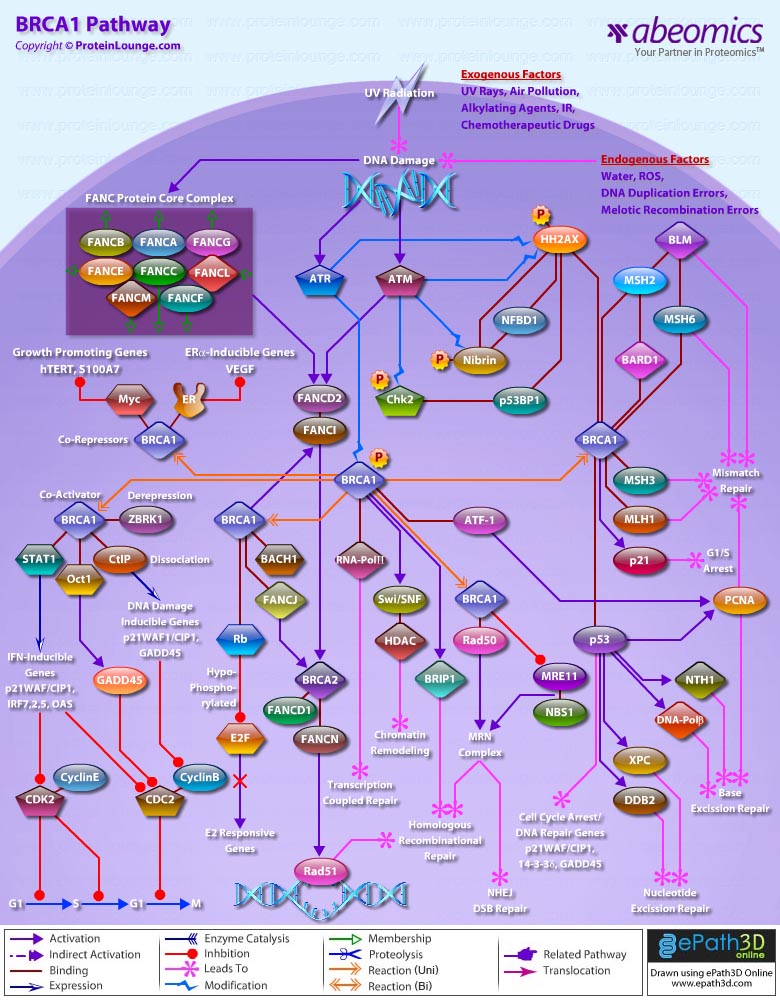
The maintenance of genome integrity is essential to all life, but is particularly important to long-lived multicellular organisms, which are susceptible to cancer. DNA damage can take the form of base modifications, strand breaks, interstrand cross-links and other lesions. To deal with many types of damage, genomes have evolved multiple cellular defense mechanisms, including DNA repair and cell cycle checkpoint processes. Different pathways exist for specific kinds of DNA damage and the cell must have ways to decide which mechanism to use for a given lesion. These requirements imply that signaling networks not only sense the presence of DNA damage, but also receive specific input such as the chemical nature of the damage, the timing of the cell cycle, the type of cell and the location of damage on the DNA. BASC (BRCA1-Associated Genome Surveillance Complex), a super complex of BRCA1 (Breast Cancer Susceptibility Protein-1), is key to recognizing and repairing DNA damage. This complex includes tumor suppressors and DNA damage repair proteins MSH2, MSH6, MLH1, ATM (Ataxia-Telangiectasia), BLM (Bloom syndrome), and the Rad50-MRE11 (Meiotic Recombination-11)-NBS1 (Nijmegen Breakage Syndrome) protein complex. In addition, RFC (DNA Replication Factor-C), a protein complex that facilitates the loading of PCNA onto DNA, is also part of BASC (Ref.1 & 2).
Eleven or more genetically distinct groups of FA have been described (FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCG, FANCE, FANCF and FANCL), each caused by recessive mutations in a different gene (Ref.4, 5 & 6). DNA damage activates the monoubiquitylation of FANCD2 (Fanconi Anemia subtype D2 protein), which is targeted to subnuclear foci, where it co-localizes with BRCA1 and Rad51 (Ref.3). FA (Fanconi Anemia) is genetically heterogeneous, with at least eleven or more genetically distinct groups (FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG and FANCL), each caused by recessive mutations in a different gene (Ref.4, 5 & 6) (Ref.3). Five of the proteins (FANCA, FANCC, FANCE, FANCF and FANCG) assemble in a multisubunit nuclear complex required for the activation of FANCD2 to a monoubiquitinated isoform (FANCD2-Ub), either in response to DNA damage or during S-Phase of the Cell Cycle, thereby targeting FANCD2 to DNA repair nuclear foci containing BRCA1, BRCA2 and Rad51, which are important in maintaining genomic stability by promoting homologous recombination repair (Ref.7). BRCA1 is a nuclear phosphoprotein, which interacts with Rad51, a human homolog of RecA, and with the Rad50-MRE11-NBS1 complex. In living cells, BRCA1 exists mostly as a heterodimeric complex with BARD1 (BRCA1-Associated RING Domain-1). BARD1 is involved in BRCA1-mediated tumor suppression. BRCA1-BARD1 co-localizes with DNA replication and repair factors in response to DNA damage. BRCA1-BARD1 heterodimers exhibit significant E3 Ub Ligase activity and the BARD1 RING finger domain greatly potentiates the Ligase activity of the BRCA1 RING finger (Ref.8). The ATM and ATR (ATM and Rad3 related) kinases, both implicated in responses to genotoxic stress, are also involved for the radiation-induced phosphorylation of BRCA1 (Ref.9). Normally, ATM phosphorylates Chk2 (Chk1 Checkpoint Homolog), which in turn phosphorylates BRCA1. The ring finger of BRCA1 confers ubiquitin ligase activity that is markedly enhanced when complexed with another ring-containing protein, BARD1 (BRCA1 Associated Ring Domain-1), and is required for the function of this tumor suppressor protein in protecting genomic integrity (Ref.10). ATR and ATM kinase targets also include repair enzymes like Rad51, Chk1 and Chk2. In response to ionizing radiation, ATM phosphorylates NBS1 leading to phosphorylation of FANCD2 and the establishment of an S-Phase checkpoint response, and in response to Mitomycin-C or Hydroxyurea, NBS1 assembles in nuclear foci with MRE11-Rad50 and FANCD2. Like ATM, the MRE11 complex is a crucial upstream regulator of checkpoint responses and DNA-repair responses in all eukaryotic cells. The MRE11 complex assembles with BRCA1 in nuclear foci following DNA damage and regulate homologous recombination repair (Ref.11). BRCA2 functions upstream in the pathway by promoting FA-complex assembly and FANCD2 activation, and/or downstream by transducing signals from FA proteins to Rad51.
BRCA1 has also been implicated in gene regulation. It induces GADD45, a p53-regulated and stress-inducible gene that plays an important role in cellular response to DNA damage. BRCA1 activation of the GADD45 promoter is mediated through the OCT1 and CAAT motifs located at the GADD45 promoter region (Ref.12). BRCA1 can trigger a G1 arrest that is mediated by transcriptional activation of p21Waf1/Cip1. In addition to its association with holoenzyme, BRCA1 can bind to several different transcription factors, including p53, Myc, STAT1, and CtIP (CBP-Interacting Protein) (Ref.13). BRCA1 acts in concert with STAT1 to differentially activate transcription of a subset of IFN-Gamma target genes and mediates growth inhibition by this cytokine. BRCA1 also binds preferentially to the hypophosphorylated form of Rb (Retinoblastoma Protein) (Ref.14). The carboxy-terminal region of the tumor suppressor protein BRCA1 is a functionally significant domain. CtIP, interacts specifically with the carboxy-terminal segment of BRCA1 from residues 1602-1863 but the exact function of CtIP is unknown. The C-terminal domain of BRCA1 (BRCT) activates transcription and interacts with RNA Polymerase holoenzyme. The BRCA1 RING finger associates with ATF1, a member of the cAMP response element-binding protein/activating transcription factor (CREB/ATF) family and leads to transcriptional activation of ATF1 target genes, some of which are involved in the transcriptional response to DNA damage (Ref.13). DNA repair by homologous recombination is mediated by the BRCA1-associated surveillance complex (comprised of BLM, MSH2–MSH6 and MRE11–Rad50–NBS1). BRCA1 can form complexes with both BACH1 and SWI/SNF to mediate chromatin remodeling and homologous recombination. HDACs regulate the access of the SWI/SNF–BRCA1 complex to DNA. Finally, BRCA1 interacts with Chk1 and PLK1 (Polo-Like Kinase-1) to regulate the G2/M and G1/S checkpoints, possibly via GADD45; thereby linking BRCA1 to the regulation of apoptosis.
BRCA1 is a tumor suppressor gene implicated in the predisposition to early onset breast and ovarian cancer. Loss of the tumor suppressor BRCA1 results in profound chromosomal instability. As a tumor suppressor, BRCA1 exerts a pleiotropic effect, playing a role in the maintenance of genomic integrity. Several functions have also been ascribed to BRCA1 including double strand DNA break repair, participating in genome surveillance, transcription-coupled DNA repair, transcriptional regulation, chromatin remodeling, and ubiquitin ligation and cell cycle checkpoint arrests. In cells, loss of BRCA1 function leads to spontaneous chromosome breakage and sensitivity to DNA damage (Ref.15).
References
1.FANCD2: a branch-point in DNA damage response?
Grompe M.
Nat Med. 2002 Jun;8(6): 555-6. No abstract available.
Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J.
Genes Dev. 2000 Apr 15;14(8):927-39.
Pang Q, Keeble W, Christianson TA, Faulkner GR, Bagby GC.
EMBO J. 2001 Aug 15; 20(16): 4478-89.
4.Biallelic inactivation of BRCA2 in Fanconi anemia.
Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D’Andrea AD.
Science. 2002 Jul 26; 297(5581): 606-9.
5.Molecular pathogenesis of Fanconi anemia: recent progress.
Taniguchi T, D’Andrea AD.
Blood. 2006 Jun 1;107(11):4223-33. Epub 2006 Feb 21.
6.Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes.
Levitus M, Rooimans MA, Steltenpool J, Cool NF, Oostra AB, Mathew CG, Hoatlin ME, Waisfisz Q, Arwert F, de Winter JP, Joenje H.
Blood. 2004 Apr 1;103(7):2498-503. Epub 2003 Nov 20.
7.Biallelic inactivation of BRCA2 in Fanconi anemia.
Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D’Andrea AD.
Science. 2002 Jul 26; 297(5581): 606-9.
8.Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein.
Xia Y, Pao GM, Chen HW, Verma IM, Hunter T.
J Biol Chem. 2003 Feb 14;278(7):5255-63. Epub 2002 Nov 12.
9.Constitutive association of BRCA1 and c-Abl and its ATM-dependent disruption after irradiation.
Foray N, Marot D, Randrianarison V, Venezia ND, Picard D, Perricaudet M, Favaudon V, Jeggo P.
Mol Cell Biol. 2002 Jun; 22(12): 4020-32.
10.Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase.
Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ.
J Biol Chem. 2002 Jun 14; 277(24): 22085-92. Epub 2002 Apr 01.
11.The Fanconi anaemia/BRCA pathway.
D’Andrea AD, Grompe M.
Nat Rev Cancer. 2003 Jan; 3(1): 23-34. Review.
12.BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs.
Fan W, Jin S, Tong T, Zhao H, Fan F, Antinore MJ, Rajasekaran B, Wu M, Zhan Q.
J Biol Chem. 2002 Mar 8;277(10):8061-7. Epub 2002 Jan 3.
13.BRCA1 physically and functionally interacts with ATF1.
Houvras Y, Benezra M, Zhang H, Manfredi JJ, Weber BL, Licht JD.
J Biol Chem. 2000 Nov 17;275(46):36230-7.
14.BRCA1-associated growth arrest is RB-dependent.
Aprelikova ON, Fang BS, Meissner EG, Cotter S, Campbell M, Kuthiala A, Bessho M, Jensen RA, Liu ET.
Proc Natl Acad Sci U S A. 1999 Oct 12;96(21):11866-71.
15.Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains.
Mallery DL, Vandenberg CJ, Hiom K.
EMBO J. 2002 Dec 16; 21(24): 6755-62.
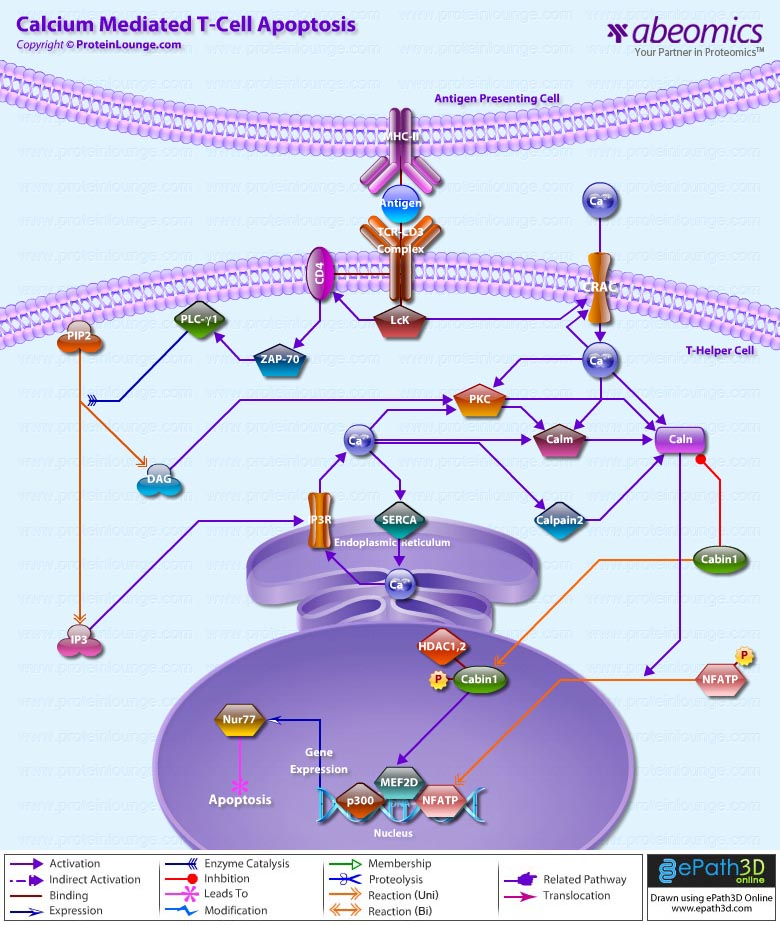
Ca2+ (Calcium) plays a major role in life and death within T-Cells. Elevation of intracellular free Ca2+ is one of the key triggering signals for T-Cell activation by antigen. The binding of antigen, MHC Class-II (Major Histocompatibility Complex Class-II), to the TCR-CD3 (T-Cell Antigen Receptor)-CD3 (CD3 Antigen) triggers the recruitment of a series of tyrosine kinases and substrates to the TCR/CD3/CD4 (CD4 Antigen) Complex, ultimately resulting in the phosphorylation and activation of PLC-Gamma1 (Phospholipase-C-Gamma1). PLC-Gamma1 cleaves PIP2 (Phosphatidylinositol 4,5-bisphosphate) in the plasma membrane to generate DAG (Diacylglycerol), which activates PKC (Protein Kinase-C) and IP3 (Inositol 1,4,5-trisphosphate), that causes entry of Ca2+ to cytosol from two sources: the ER (Endoplasmic Reticulum) and the extracellular space. IP3 generated subsequent to TCR-CD3 stimulation binds to receptors in the ER membrane, opening Ca2+ channels that release Ca2+ to the cytosol (Ref.1). Release is a highly cooperative process, owing to both the binding of multiple IP3 molecules to the tetrameric receptor and to positive feedback by Ca2+ released from the ER. The activity of the IP3R/ITPR (Inositol 1, 4, 5-Triphosphate Receptor) is increased during the early phase of T-Cell activation by Fyn (Fyn Oncogene Related to Src, FGR, YES). IP3R release Ca2+ from intracellular stores and by depleting the stores, trigger prolonged Ca2+ influx through CRAC (Ca2+ Release-Activated Ca2+ Channel/Capacitance-Regulated Activation Channel/Store-Operated Calcium Release-Activated Ca2+/Ca2+ Release-Activated Ca2+ Current) in the plasma membrane. There are distinguished different classes of SOCs (Store-Operated Channels); the class found in T-Cells is the CRAC channel, named for a similar channel originally found in mast cells. It is distinguished from other SOCs primarily by its extremely high Ca2+ selectivity. At short times, Ca2+ signals help to stabilize contacts between T-Cells and Antigen-Presenting Cells through changes in motility and cytoskeletal reorganization (Ref.2).
Mechanisms that remove Ca2+ from the cytosol also exert a powerful influence over the amplitude, duration and dynamics of the net Ca2+ signal. SERCA (Sarco-Endoplasmic Reticulum Ca2+-ATPases) pumps Ca2+ from the cytosol into the ER. SERCA form a significant part of the Ca2+ signaling network in T-Cells by accumulating Ca2+ in the ER, thereby enabling Ca2+ transients via IP3-induced release and controlling store-operated Ca2+ entry. Changes in Ca2+ are associated with a decrease in accumulation of NFATP (Nuclear Factor of Activated T-Cells Pre-existing Component) in the nucleus. NFATP has higher Ca2+ sensitivity but is rapidly reversible. The best current candidates for SOC genes are mammalian homologs of the Drosophila TRP (Transient Receptor Potential) gene. The lack of Ca2+ selectivity or insensitivity to store depletion does not necessarily rule out a role for any particular human TRP in helping to make an endogenous CRAC channel (Ref.3). Focusing on CRAC, which is the only reliably SOC described so far, the crucial experiment will be whether the correct pore properties of CRAC will emerge in a heteromer or a signalplex. This has not yet been shown. There is no doubt that multimerisation occurs for many TRPs, such as TRPC1 (Transient Receptor Potential Cation Channel Subfamily-C Member-1)/4/5, TRPC3/6/7, TRPV1 (Transient Receptor Potential Cation Channel Subfamily-V Member-1)/3, TRPV5/6, TRPM4 (Transient Receptor Potential Cation Channel Subfamily-M Member-4)/5, TRPM6/7 and that heteromer formation changes permeation and kinetic properties of those channels. But so far, no CRAC channel seems to be present in the TRP family since most TRP channels have a low Ca2+ selectivity. In any case, the superficial identification of TRPs as SOCs must be avoided. Then the question remains, what else, if not TRPs? What is the message that links store depletion to channel activation? Although it is too early to say whether CIF (Calcium Influx Factor) directly activates CRAC channels, CIF accelerates CRAC activation relative to activation by EGTA (Ethylene Glycol bis(â-aminoethylether)- N,N,N’,N’-Tetraacetic Acid/ Ethylenebis (Oxyethylenenitrilo) Tetraacetic Acid) alone (Ref.1 & 4).
The “Inside-Out Signaling Pathway” that causes the influx of Ca2+ through specialized CRAC channels provides the persistent Ca2+ signal necessary to maintain NFATP proteins in the nucleus (Ref.2). The major Ca2+ and Caln (Calcineurin)-responsive elements in the Nur77 (Mouse Homolog of Nur77) promoter are binding sites for MEF2D (MADS Box Transcription Enhancer Factor-2 Polypeptide-D). NFATP interacts with MEF2D and enhances its transcriptional activity, offering a plausible mechanism of activation of MEF2D by Caln. NFATP synergizes with MEF2D to recruit the co-activator p300 for the transcription of Nur77. Surprisingly, the enhancement of transcriptional activity of MEF2D by NFATP does not require its DNA-binding activity, proving that NFATP acts as a co-activator for MEF2D. Transient co-expression of p300, MEF2D, NFATP and constitutively active Caln is sufficient to recapitulate TCR signaling for the selective induction of the endogenous Nur77 gene. These results implicate NFATP as an important mediator of T-Cell apoptosis (Ref.5). Binding of Cabin1 (Calcineurin Binding Protein-1) to MEF2D suppresses MEF2D transcriptional activity. However, in the presence of a Ca2+ signal, Calm (Calmodulin) binds to Cabin1, freeing MEF2D to recruit the co-activator p300 for transcriptional activation of MEF2D target genes. Cabin1-MEF2 interaction is required for proper MEF2D induction and phosphorylation after TCR signaling. The COOH-terminal region of Cabin1 interacts with MEF2D and Calm in a mutually exclusive manner. The interaction between Cabin1 and Caln is dependent on both Ca2+ signal and PKC activation, which results in Cabin1 hyperphosphorylation. As Cabin1 is occurs primarily in the nucleus in T-Cells, it interacts only with activated Caln that has translocated into the nucleus (Ref.6).
Capn2 (Calpain-2) cleaves the Caln-binding domain of Cabin1 to activate Caln and elicit Ca2+-triggered cell death. Cabin1 cleavage and Caln activation are suppressed by Capn2 inhibitors. The cleavage of Cabin1 allows Cabin1 to be inactivated and dissociated from the Caln complex, leading to the activation of Caln (Ref.7). In unactivated T-Cells, MEF2D is bound to a transcriptional repression complex consisting of Cabin1, HDAC1 (Histone Deacetylase-1) and HDAC2. Upon TCR signaling and Ca2+ influx, activated Calm binds to Cabin1, releasing it from MEF2D, vacating the MADS/MEF2 domain for association with the coactivator p300. The Ca2+-dependent association and dissociation of two opposing classes of chromatin remodeling enzymes are responsible for tight control of Nur77 transcription, ensuring that thymocytes will not commit to apoptosis in the absence of TCR signaling. Cabin1 also competes with p300 for binding to MEF2D. Activation of MEF2D and the consequent transcription of Nur77 are controlled by the association of MEF2D with the HDACs via the Ca2+-dependent repressor Cabin1 (Ref.8). The role of Ca2+ signaling in regulating MEF2D-dependent gene expression in non-lymphoid tissues still remains the subject of ongoing investigation (Ref.9).
References
1.Calcium signaling mechanisms in T lymphocytes.
Lewis RS.
Annu. Rev. Immunol. 2001;19:497-521.
2.NFAT signaling: choreographing the social lives of cells.
Crabtree GR, Olson EN.
Cell. 2002 Apr;109 Suppl:S67-79.
3.TRP ion channels in the nervous system.
Moran MM, Xu H, Clapham DE.
Curr. Opin. Neurobiol. 2004 Jun;14(3):362-9.
Wilk S.
Sci. STKE. 2005 May 24;2005(285):tr16.
5.Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis.
Youn HD, Chatila TA, Liu JO.
EMBO J. 2000 Aug 15;19(16):4323-31.
Esau C, Boes M, Youn HD, Tatterson L, Liu JO, Chen J.
J. Exp. Med. 2001 Nov 19;194(10):1449-59.
Kim MJ, Jo DG, Hong GS, Kim BJ, Lai M, Cho DH, Kim KW, Bandyopadhyay A, Hong YM, Kim do H, Cho C, Liu JO, Snyder SH, Jung YK.
Proc. Natl. Acad. Sci. U S A. 2002 Jul 23;99(15):9870-5.
Youn HD, Liu JO.
Immunity. 2000 Jul;13(1):85-94.
9.Ca(2+)-dependent gene expression mediated by MEF2 transcription factors.
Blaeser F, Ho N, Prywes R, Chatila TA.
J. Biol. Chem. 2000 Jan 7;275(1):197-209.
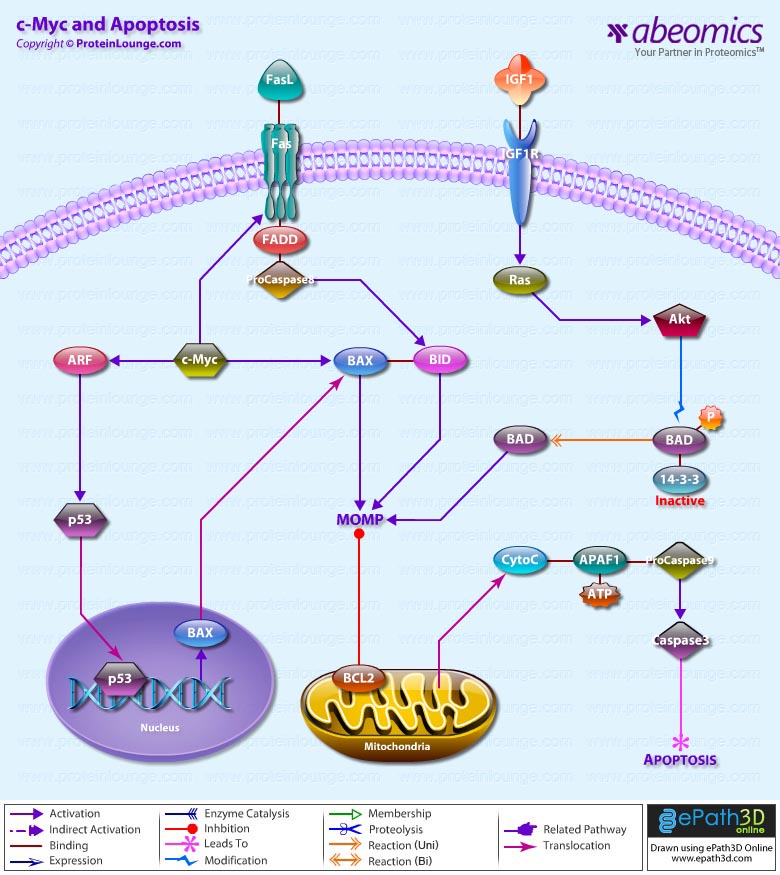
Many proto-oncogenes participate in the regulation of apoptosis and closely intertwined with their actions are various growth factors and other genes that participate in the control of cellular growth. The proto-oncogene c-Myc encodes a transcription factor c-Myc that plays a critical role in multiple cellular processes including cell growth, proliferation, differentiation, and apoptosis. Expression of c-Myc is rapidly induced by mitogens and is down regulated during differentiation. c-Myc activity is sufficient to drive cells into the cell cycle in the absence of growth factors but also induces apoptosis when survival factors are missing. Deregulated expression of c-Myc is associated with a wide array of human cancers. Thr-58 is a major phosphorylation site in c-Myc and is a mutational hotspot in Burkitt’s and other aggressive human lymphomas. The quantity of c-Myc is carefully controlled by many mechanisms, and it exerts its oncogenic effects through regulation of genes involved with growth and proliferation (Ref.1).
Expression of c-Myc sensitizes cells to a wide range of mechanistically diverse proapoptotic insults, including DNA damage, death receptor signaling, hypoxia, genotoxic stress, and nutrient deprivation. Two discrete proapoptotic effector pathways mediate this sensitization. One of these involves stabilization of p53 through the ARF (Active Response Factor)/MDM2 (Mouse Double Minute-2) pathway, which serves as a sentinel for genotoxic damage. The second promotes release of Cyto C (Cytochrome-C) from mitochondria into the cytosol, possibly through activation of the pro-apoptotic molecule BAX (BCL2 Associated-X Protein) by a mechanism that is independent of both the Fas-FasL and the DNA damage proapoptotic pathways (Ref.2). Activated BAX within the mitochondrial membrane leads to creation or alteration of membrane pores, resulting in MOMP (Mitochondrial-Outer-Membrane Permeabilization). Once released into the cytosol, Cyto C associates with APAF1 (Apoptotic Protease Activating Factor-1) protein and procaspase-9 to form the apoptosome (‘wheel of death’). In the presence of ATP, Caspase 9 is activated, leading to activation of downstream effector caspases, including Caspase 3, which ultimately leads to the degradation of cell components and the demise of the cell. Since the release of Cyto C is the principal target for suppression by survival factors, this pathway acts as a trophic sentinel (Ref.3), triggering Myc-induced apoptosis. c-Myc-induced release of Cyto C is also suppressed by BCL2 (B-Cell CLL/Lymphoma-2)/BCL-XL, which, like survival factors, potently exacerbates c-Myc oncogenicity. Both of these apoptotic pathways share APAF1 and Caspase 9 as final apoptotic effectors downstream of the mitochondrion. Inhibition of this mitochondrial pathway, either by suppression of Cyto C release by survival signals or BCL2/BCL-XL proteins or by incapacity of the downstream mitochondrial apoptotic effector pathway through genetic loss of APAF1 or Caspase 9 (Ref.4) that inhibits c-Myc-induced apoptosis and promotes c-Myc oncogenicity.
Ectopic expression of the death receptor signaling proteins or ligation of the death receptor CD95/Fas triggers the association of the intracellular adaptor protein FADD (FAS-associated death domain), which then recruits procaspase-8, resulting in its auto-activation. Caspase-8 also activates the pro-apoptotic protein BID (BH3 Interacting domain Death agonist), which promotes MOMP. Survival signals that serve to block c-Myc-induced apoptosis include signaling via the IGF1R (Insulin-like Growth Factor-1 Receptor) or activated Ras, which leads to the activation of Akt1 serine/threonine kinase and subsequent phosphorylation of the pro-apoptotic protein BAD (BCL2 Associated Death Promoter). Phosphorylated BAD is sequestered and inactivated by cytosolic 14-3-3 proteins. Anti-apoptotic proteins, BCL2 and BCL-XL also block Cyto C release, possibly through the sequestration of BAX (Ref.5).
The oncogene c-Myc is frequently associated with human malignancies and cancer-related mutations in c-Myc lead to defects in its degradation and thereby contribute to the increase in its cellular level that is associated with the disease. Recent studies suggest that c-Myc is able to activate the cell cycle machinery and intriguingly, its ability to activate glycolysis suggests that in addition to triggering the cell cycle, c-Myc also sustains the fuel necessary to run the cell cycle machinery. Indeed, its ability to enhance the activities of specific enzymes involved in DNA metabolism and other metabolic pathways further suggests that it is a key molecular integrator of cell cycle machinery and cellular metabolism (Ref.6).
References
1.MYC oncogenes and human neoplastic disease.
Nesbit CE, Tersak JM, Prochownik EV.
Oncogene. 1999 May 13; 18(19): 3004-16. Review.
2.c-Myc-induced sensitization to apoptosis is mediated through Cytochrome c release.
Juin P, Hueber AO, Littlewood T, Evan G.
Genes Dev. 1999 Jun 1; 13(11): 1367-81.
3.Proliferation, cell cycle and apoptosis in cancer.
Evan GI, Vousden KH.
Nature. 2001 May 17; 411(6835): 342-8. Review.
4.Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition.
Soengas MS, Alarcon RM, Yoshida H, Giaccia AJ, Hakem R, Mak TW, Lowe SW.
Science. 1999 Apr 2; 284(5411): 156-9.
Klefstrom J, Verschuren EW, Evan G.
J Biol Chem. 2002 Nov 8; 277(45): 43224-32. Epub 2002 Aug 28.
6.c-Myc target genes involved in cell growth, apoptosis, and metabolism.
Dang CV.
Mol Cell Biol. 1999 Jan; 19(1): 1-11. Review.
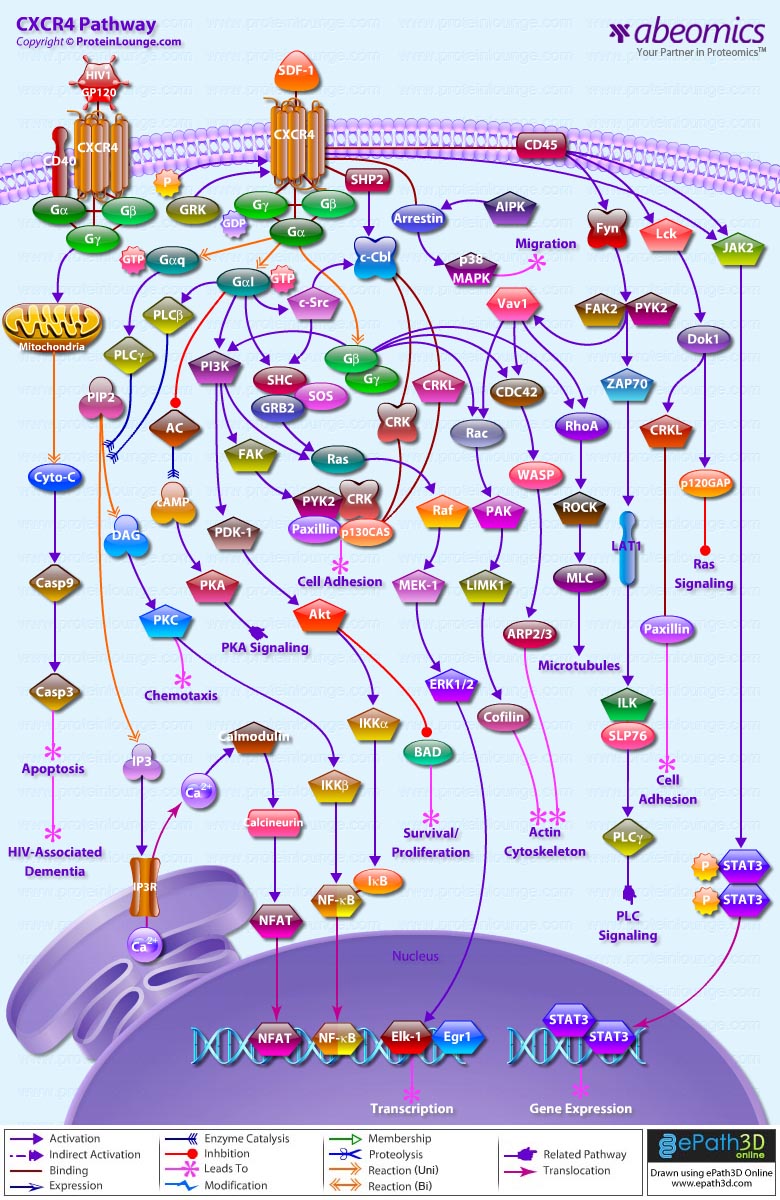
The Chemokine receptor CXCR4 is a 352 amino acid rhodopsin-like GPCR and selectively binds the CXC chemokine Stromal Cell-Derived Factor 1 (SDF-1) also known as CXCL12. Classically, two alternatively spliced isoforms of SDF have been identified. SDF-1Alpha is an 89 amino acid protein that is the predominantly expressed form of SDF-1 while SDF-1Beta contains a four amino acid extension at the carboxyl terminus. Recently, an additional four splice variants that contain 30 (SDF-1Gamma), 31 (SDF-1Delta), 1 (SDF-1Epsilon), and 51 (SDF-1phi) amino acid extensions at the carboxyl terminus compared to SDF-1Alpha have been identified (Ref.1).
CXCL12 is the sole ligand for the chemokine receptor CXCR4.This receptor has been described as undergoing dimerization after binding to CXCL12 or alternately in its unbound configuration. Upon binding of CXCL12 to CXCR4, the receptor is stabilized into a conformation that activates the heterotrimeric G-protein (G-alpha and G-beta gamma), G-alphaI and G-AlphaQ being the major components. G-alphaQ proteins activate phosphatidylinositol-specific phospholipases such as Phospholipase C-gamma (PLC-gamma), which hydrolyzes Phosphatidylinositol-4,5-biphosphate (PIP2) to generate Inositol triphosphate (IP3) and Diacylglyerol (DAG). These events lead to calcium flux and the activation of several PKC isoforms that have been shown to be important for SDF-1-induced chemotaxis. G-alphaI activates Phospholipases, phosphodiesterases, and the lipid kinase PI3K via Src-family kinases.Signaling through PI3K pathway leads to the activation of PAK and cell polarization, the first step in migration. PI3K and various tyrosine kinases that activate Akt and CDC42 are involved in actin polymerization. PLC¨Cmediated events, such as calcium release and protein kinase C activation, as well as focal adhesion kinase, Pyk2, Paxillin, and ERK are important in the adhesion process, leading to cell migration. In parallel, activation of Akt, ERK, and tyrosine kinases such as Src leads to transcription of genes involved in migration.G-alphaI activation inhibits adenylyl cyclases that in turn lead to a reduction in cAMP levels as well as the activation of phospholipases and phosphodiesterases (Ref.2 and 3).
G-beta gamma also activates PI3K. PI3K activation stimulates downstream targets such as protein kinase B (PKB/Akt), NF-kappaB, mitogen/extracellular signal-regulated kinase (MEK-1), and extracellular signal-regulated kinase (ERK1/2). PI3K also triggers the tyrosine phosphorylation of focal adhesion complex components such as proline-rich tyrosine kinase (Pyk2), Paxillin, Crk, and p130Cas. GTP-bound G-betagama stimulates guanine nucleotide exchange factors (GEFs) such as TIAM1 and PREX1 specific for the Rho family GTPases (Rac/CDC42/RhoA). These GTPases activate pathways regulating cytoskeleton: Rac activates p21-activated kinase (PAK), which then activates LIM kinase (LIMK), leading to cofilin phosporylation and actin polymerization. CDC42 promotes actin assembly through the Wiskott-Aldrich Syndrome family protein (WASP) and actin-nucleating protein ARP2/3.RhoA activates Rho kinase (ROCK) , leading to myosin light-chain (MLC) phosporylation and microtubule rearrangement. Janus-activated kinase/ signal transducers and activators of transcription (JAK/STAT) pathway is activated through CXCR4, partially independent of G-protein. The association of JAK with CXCR4 activates STAT proteins, which translocate to the nucleus and regulate transcription (Ref.2 and 3).
Down-regulation of the CXCR4 receptor is initiated by phosphorylation of its cytoplasmic tail. Subsequent to the binding of arrestin to the COOH terminus of CXCR4, the receptor is internalized through endocytosis. Degradation of CXCR4 occurs in the lysosome. The CXCR4¡¤CXCR7 heterodimer complex recruits Beta-arrestin, resulting in preferential activation of Beta-arrestin-linked signaling pathways over canonical G protein pathways, including ERK1/2, p38 MAPK, and SAPK. Stimulation of other G-protein coupled receptors may also down-regulate CXCR4 signaling through a process called heterologous desensitization (Ref. 3 and 4).
CXCR4 also acts as cell surface coreceptors of HIV-1 strains forming heterodimers with CD4, the principal HIV-1 receptor. On the virion surface, the viral envelope is arranged in spike©like structures formed by trimeric complexes of gp120, the external subunit that mediates virion attachment, and gp41, the transmembrane subunit that mediates the fusion process. For HIV-1 to enter a target cell, the viral envelope glycoprotein gp120 must interact with CD4 and CXCR4. This two©stage receptor©interaction strategy allows HIV©1 to maintain the highly conserved coreceptor©binding surface in a cryptic conformation, unraveling it only upon binding of gp120 to CD4. Binding of the HIV-1 envelope to its chemokine coreceptors mediates two major biological events: membrane fusion and signaling transduction (Ref.5 and 6). The CXCL12/CXCR4 pathway is a target for therapeutics that block CXCL12/CXCR4 interaction or inhibit downstream intracellular enzyme activities. Small molecular inhibitors of CXCR4, such as plerixafor or BKT140, and blocking antibodies toward CXCR4 or CXCL12 are being investigated in various cancer settings. CXCL12/CXCR4 disruption is essential for the egress of hematopoietic stem and/or progenitor cells from bone marrow into circulation. Conversely, CXCL12/CXCR4 function is essential for homing and/or engraftment of hematopoietic stem cells to the bone marrow after transplantation (Ref. 7).
References
1.Regulation of CXCR4 signaling.
Busillo JM, Benovic JL.
Biochim Biophys Acta. 2007 Apr;1768(4):952-63. Epub 2006 Nov 10. Review.
2.Translating an Antagonist of Chemokine Receptor CXCR4: from bench to bedside.
Wong D, Korz W.
Clin Cancer Res. 2008 Dec 15;14(24):7975-80. doi: 10.1158/1078-0432.CCR-07-4846. Review.
3.Chemokine coreceptor signaling in HIV-1 infection and pathogenesis.
Wu Y, Yoder A.
PLoS Pathog. 2009 Dec;5(12):e1000520. doi: 10.1371/journal.ppat.1000520. Epub 2009 Dec 24. Review.
4.CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration.
D¨¦caillot FM, Kazmi MA, Lin Y, Ray-Saha S, Sakmar TP, Sachdev P.
J Biol Chem. 2011 Sep 16;286(37):32188-97. doi: 10.1074/jbc.M111.277038. Epub 2011 Jul 5.
5.CCR5/CD4/CXCR4 oligomerization prevents HIV-1 gp120IIIB binding to the cell surface.
Mart¨ªnez-Muñoz L, Barroso R, Dyrhaug SY, Navarro G, Lucas P, Soriano SF, Vega B, Costas C, Muñoz-Fern¨¢ndez MÁ, Santiago C, Rodr¨ªguez Frade JM, Franco R, Mellado M.
Proc Natl Acad Sci U S A. 2014 May 13;111(19):E1960-9. doi: 10.1073/pnas.1322887111. Epub 2014 Apr 28
6.HIV and the chemokine system: 10 years later.
Lusso P.
EMBO J. 2006 Feb 8;25(3):447-56. Epub 2006 Jan 26. Review.
7.CXCL12 (SDF-1)/CXCR4 pathway in cancer.
Teicher BA, Fricker SP.
Clin Cancer Res. 2010 Jun 1;16(11):2927-31. doi: 10.1158/1078-0432.CCR-09-2329. Epub 2010 May 18. Review.
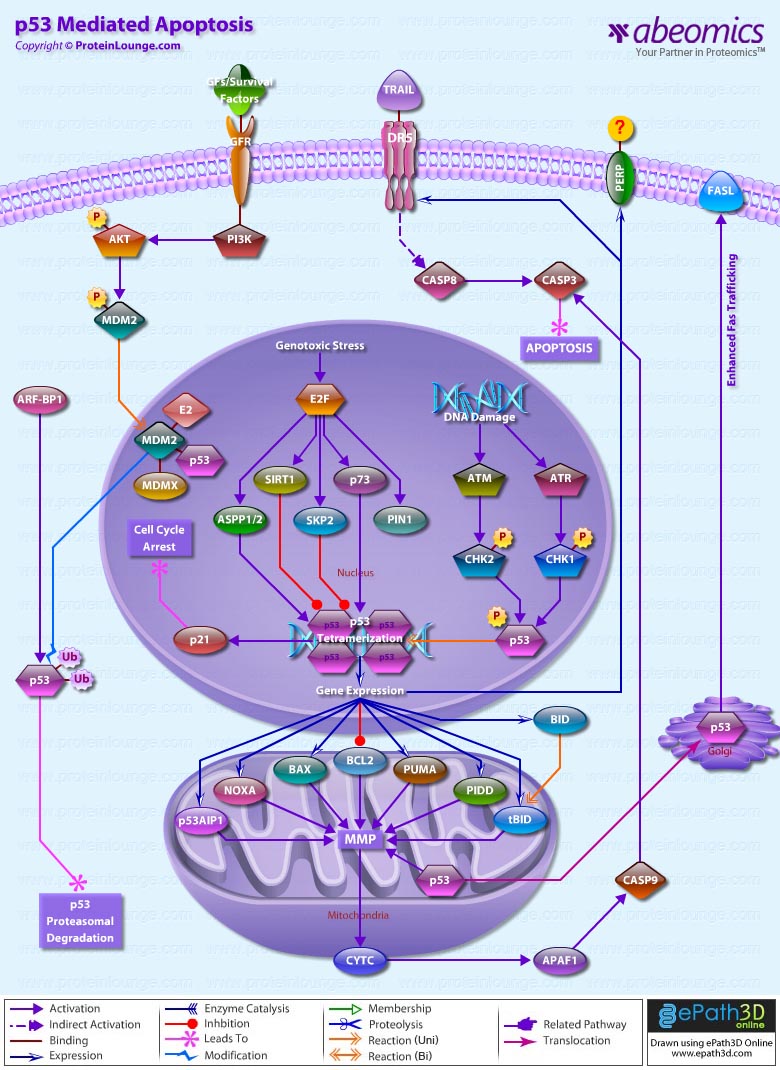
Apoptosis is a genetically controlled mechanism of cell death that is essential for the elimination of unwanted cells during normal development and for the maintenance of tissue homeostasis. One of the major apoptosis signaling pathways involves the p53 tumour suppressor. Tumor protein p53 is a nuclear transcription factor that regulates the expression of a wide variety of genes involved in apoptosis, growth arrest or senescence in response to genotoxic or cellular stress. There are four conserved domains in p53: N-terminal domain, which is required for transcriptional transactivation, a sequence-specific DNA binding domain, a tetramerization domain near the C-terminal end and a C-terminal domain that interacts directly with single stranded DNA. Having a short half-life, p53 is normally maintained at low levels in unstressed mammalian cells by continuous ubiquitylation and subsequent degradation by the 26S Proteasome. Nonphosphorylated p53 is ubiquitylated by the MDM2 (Mouse Double Minute-2) ubiquitin ligase. When the cell is confronted with stress like DNA damage, Hypoxia, Cytokines, metabolic changes, Viral infection, or Oncogenes, however, p53 ubiquitylation is suppressed and p53 is stabilized and accumulates in the nucleus. The N-terminus of p53 may be heavily phosphorylated, whereas the C-terminus may be phosphorylated, acetylated or sumoylated. The ability of p53 to elicit diverse regulatory functions is likely to depend on its phosphorylation pattern, which is conformation dependent. p53 phosphorylation is mediated by several cellular kinases including Chks (Checkpoint Kinases), CSNK1-Delta (Casein Kinase-1-Delta), CSNK2 (Casein Kinase-2), PKA (Protein Kinase A), CDK7 (Cyclin-Dependent Kinase-7), DNA-PK (DNA-Activated- Protein Kinase), HIPK2 (Homeodomain-Interacting Protein Kinase-2) and JNK (Jun NH2-terminal kinase). The main Kinases involved in p53 phosphorylation include Chk1 (Cell Cycle Checkpoint Kinase-1) and Chk2 (Cell Cycle Checkpoint Kinase-2). In response to DNA Damage Chk1 and Chk2, activated by ATR (Ataxia-Telangiectasia and Rad3 Related) and ATM Ataxia Telangiectasia Mutated Gene) respectively, phosphorylates p53. Chk2 can phosphorylate p53 on Ser20, which prevents MDM2 binding and results in p53 stabilization. ATM can also phosphorylate p53 on Ser15, which is required for activation of p53 as a transcription factor and may act synergistically with Ser20 phosphorylation. Once activated p53 binds, as a tetramer, to a p53-binding site. In doing so, it activates expression of downstream genes leading to programmed cell death and/or cell cycle arrest, thus functioning as a tumor suppressor. p53 promotes apoptosis through multiple mechanisms, including transactivation of specific target genes, down-regulation of a distinct set of genes, and transcription-independent mechanisms (Ref. 1 & 2).
p53 stimulates a wide network of signals that act through two major apoptotic pathways: Extrinsic Pathways and Intrinsic Pathways. The Extrinsic pathway involves engagement of particular `death’ receptors that belong to the tumor TNFR (Tumor Necrosis Factor Receptor) family and, through the formation of the DISC (Death-Inducing-Signaling-Complex), leads to a cascade of activation of Caspases, including Caspase8 and Caspase3, which in turn induce apoptosis. Most common death receptors involved in extrinsic apoptosis Fas, DR5 (Death Receptor-5) and PERP. The cell-surface receptor Fas (CD95/Apo-1), a member of the TNFR family of receptors, is a key component of the extrinsic death pathway. Fas is activated by binding of its ligand, FasL, which is expressed predominantly by T-Cells. p53 induces Fas mRNA expression by binding to elements found in the promoter and first intron of the Fas gene. This induction occurs in response to Gamma-irradiation, and it appears to be strictly tissue specific. In addition to stimulating Fas transcription, overexpressed p53 may enhance levels of Fas at the cell surface by promoting trafficking of the Fas receptor from the Golgi. This may allow p53 to rapidly sensitize cells to Fas-induced apoptosis before the transcription-dependent effect operates. How p53 promotes Fas trafficking is not understood (Ref. 3 & 4). The second member of this receptor family that is induced by p53 is DR5/Killer, the death-domain-containing receptor for TRAIL (TNF-Related Apoptosis-Inducing Ligand). DR5 is induced by p53 in response to DNA damage and in turn promotes cell death through Caspase8. DR5 induction is cell type specific. Another apoptotic gene, PERP, is induced in response to DNA-damage. PERP is a putative tetraspan transmembrane protein that represents a new member of the PMP-22/Gas family of proteins implicated in cell growth regulation. The kinetics of PERP induction in response to DNA damage and the presence of a p53-responsive element in the PERP promoter support the notion that it is a direct p53 target. The mechanism by which PERP contributes to p53-mediated apoptosis is yet to be defined. Perhaps analogously to Killer and Fas, PERP could serve as a cell death receptor, albeit a different type with four membrane-spanning domains, to receive either autocrine or paracrine signals. Alternatively, a sequence similarity between PERP and the calcium channel suggests that it could have channel or pore activity, perhaps allowing some crucial molecule important for activating apoptosis to pass through. However it exerts its function, PERP might ultimately act by affecting regulators of the apoptotic machinery, such as BAX (Bcl2 Associated-X Protein) or Bcl2, or by directly activating apoptotic effectors like Caspases (Ref. 3, 5 & 6).
The Intrinsic apoptotic pathway is dominated by the Bcl2 family of proteins, which governs the release of CytoC (Cytochrome-C) from the mitochondria. The Bcl2 family comprises anti-apoptotic (pro-survival) and pro-apoptotic members. Family members are classified on the basis of structural similarity to the BH (Bcl2 Homology) domains (BH1, BH2, BH3 and BH4), and a transmembrane domain. The BH3 domain, which is present in all members and is essential for heterodimerization among members, is the minimum domain required for the proapoptotic function. Pro-survival proteins members are most structurally similar to Bcl2, such as BclXL; pro-apoptotic proteins, BAX and BAK (Bcl2 Antagonist Killer-1) are structurally similar to Bcl2 and BclXL and antagonize their pro-survival functions; and the pro-apoptotic `BH3-only’ proteins. Intriguingly, a key subset of the Bcl2 family genes are p53 targets, including BAX, Noxa, PUMA (p53-Upregulated Modulator of Apoptosis) and the most recently identified, BID (BH3 Interacting Domain Death Agonist). BAX was the first member of this group shown to be induced by p53, but p53-responsive elements have only recently been unequivocally identified in the BAX gene. In response to stress activation, BAX forms a homodimer and releases CytoC from the mitochondria. CytoC, APAF1 (Apoptotic Protease-Activating Factor-1) and Procaspase9 then form a complex termed the apoptosome, in which Caspase9 is activated and promotes activation of Caspase3, Caspase6 and Caspase7, that after cleavage of vital death substrates induce the final demise of the cell. The requirement for BAX in p53-mediated apoptosis appears to be cell-type dependent (Ref. 7).
The PUMA gene is also directly induced by p53 in response to DNA damage, through p53-responsive elements within the first intron of PUMA. In humans, PUMA encodes two BH3-domain-containing proteins, PUMA-Alpha and PUMA-Beta. BAX is absolutely required for PUMA-mediated apoptosis. PUMA expression promotes mitochondrial translocation and mulitmerization of BAX, culminating in apoptosis induction. BAX thus participates in the death response as an indirect target of p53 through PUMA. Another p53 target gene, Noxa, contains a single p53-responsive element in its promoter and is induced in response to X-ray irradiation. Noxa encodes a BH3-only protein and hence is likely to contribute to p53-mediated apoptosis in a similar manner to PUMA and BAX, although this is yet to be demonstrated. Thus, in response to DNA damage, p53 activates the intrinsic mitochondrial apoptotic pathway by inducing the expression of at least three Bcl2 pro-apoptotic family members, shifting the balance towards pro-apoptotic effects. An important p53 transcription target gene was identified as p53AIP1 (p53-regulated apoptosis-inducing protein 1), which is located in the mitochondrial membrane and is directly involved in p53-dependent mitochondrial apoptosis. Recent evidence has suggested the existence of a transcription-independent pathway of p53-mediated apoptosis. A fraction of the p53 molecules that accumulate in damaged cells translocates to mitochondria, and this translocation is sufficient for p53 to induce permeabilization of the outer mitochondrial membrane through formation of complexes with the protective proteins BclxL and Bcl2, resulting in the release of CytoC into the cytosol. p53 has been shown to transcriptionally repress the antiapoptotic genes Bcl2, BclxL and Survivin (Ref. 8, 9 & 10).
Although Extrinsic and Intrinsic pathways are two separate pathways, recent studies link the Extrinsic and Intrinsic pathways, lending support to the idea of converging rather than distinct pathways. The pro-apoptotic BID connects the activation of the extrinsic death receptor pathway to activation of the mitochondrial-disruption processes associated with the intrinsic pathway. Activation of BID involves cleavage of cytoplasmic BID by Caspase8 to expose a new N-terminal glycine residue, which undergoes post-translational myristoylation. Myristoylated BID translocates to the mitochondria, inserts into the membrane and activates BAX and BAK to initiate mitochondrial events leading to apoptosome formation. The BID gene is transcriptionally regulated by p53 in response to Gamma-irradiation through response elements in the first intron of the human gene. p53 therefore appears to promote the convergence of the intrinsic and extrinsic pathways through BID regulation (Ref.11). Akt also play an important role in regulating p53 pathway under growth/survival conditions and under stress signals. Under normal conditions, activation of Akt can begin with several events, mainly the binding of a Ligand to a Receptor in the cell membrane. Most common Ligands activating Akt include Growth factors, Cytokines, Mitogens and Hormones. Growth factors bind to RTK (Receptor Tyrosine Kinase) and cause autophosphorylation of tyrosine residues on the intracellular domain of the receptor. PI3K (Phosphoinositol 3-Kinase) is recruited to the phosphotyrosine residues (consensus sequence pYXXM) via SH2 domains in the regulatory domain (p85), and is therefore targeted to the inner cell membrane. Binding of the p85 subunit of PI3K to the phosphorylated RTK leads to conformational changes in the catalytic domain of PI3K (p110) and consequent kinase activation. PI3K then phosphorylates membrane bound PIP2 (Phosphatidylinositol-3,4-Bisphosphate) to generate PIP3 (Phosphatidylinositol-3,4,5-Triphosphate). The binding of PIP3 to the PH domain anchors Akt to the plasma membrane and allows its phosphorylation and activation by PDK1 (Phosphoinositide-Dependent Kinase-1). The negative regulation of p53 by AKT is induced in response to survival signals from MDM2. The activation of this pathway leads to the inhibition and destruction of p53. Under stress conditions this pathways is blocked through the cleavage and degradation of Akt, and the inhibition of PI3K through PTEN (Phosphatase and Tensin Homolog). Both of these activities are induced by p53 (Ref. 12 & 13).
Thus, a multitude of mechanisms are employed by p53 to ensure efficient induction of apoptosis in a stage-, tissue- and stress-signal-specific manner. Mutations in p53 are associated with genomic instability and increased susceptibility to cancer. It is the most frequently mutated protein in all cancer with an estimated 60% of all cancers having mutated forms that affect its growth suppressing activities. However some common tumours have a higher incidence such that 90% of cervical and 70% of colorectal are found to have p53 mutations. The p53 protein can be inactivated in several ways, including inherited mutations that result in a higher incidence of certain familial cancers such as Li-Fraumeni syndrome. Certain DNA tumour viruses, such as the human adenovirus and the papilomavirus, bind to and inactivate the protein. Functional p53 is thought to provide a protective effect against tumorigenesis (Ref.14 & 15).
References
1.The role of p53 in apoptosis.
Amaral JD, Xavier JM, Steer CJ, Rodrigues CM.
Discov Med. 2010 Feb;9(45):145-52.
2.p53 Family: Role of Protein Isoforms in Human Cancer.
Wei J, Zaika E, Zaika A.
J Nucleic Acids. 2012;2012:687359. Epub 2011 Oct 9.
3.Placental apoptosis in health and disease.
Sharp AN, Heazell AE, Crocker IP, Mor G.
Am J Reprod Immunol. 2010 Sep;64(3):159-69.
4.p53-mediated delayed NF-?B activity enhances etoposide-induced cell death in medulloblastoma.
Meley D, Spiller DG, White MR, McDowell H, Pizer B, Sée V.
Cell Death Dis. 2010 May 13;1:e41.
5.p53 and the regulation of hepatocyte apoptosis: implications for disease pathogenesis.
Amaral JD, Castro RE, Steer CJ, Rodrigues CM.
Trends Mol Med. 2009 Nov;15(11):531-41. Epub 2009 Oct 12.
Davies L, Spiller D, White MR, Grierson I, Paraoan L.
Cell Death Dis. 2011 Mar 31;2:e136.
Zhang XP, Liu F, Wang W.
J Biol Chem. 2010 Oct 8;285(41):31571-80. Epub 2010 Aug 4.
8.PUMA, a potent killer with or without p53.
Yu J, Zhang L.
Oncogene. 2008 Dec;27 Suppl 1:S71-83.
9.Noxa: at the tip of the balance between life and death.
Ploner C, Kofler R, Villunger A.
Oncogene. 2008 Dec;27 Suppl 1:S84-92.
10.p53: the attractive tumor suppressor in the cancer research field.
Ozaki T, Nakagawara A.
J Biomed Biotechnol. 2011;2011:603925. Epub 2010 Dec 6.
11.Post-translational myristoylation: Fat matters in cellular life and death.
Martin DD, Beauchamp E, Berthiaume LG.
Biochimie. 2011 Jan;93(1):18-31. Epub 2010 Nov 5.
12.Oscillations of the p53-Akt network: implications on cell survival and death.
Wee KB, Surana U, Aguda BD.
PLoS One. 2009;4(2):e4407. Epub 2009 Feb 6.
13.Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents.
Degtyarev M, De Mazière A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, Davis DP, Stern HM, Murray LJ, Hoeflich KP, Klumperman J, Friedman LS, Lin K.
J Cell Biol. 2008 Oct 6;183(1):101-16.
14.Mutant p53 gain-of-function in cancer.
Oren M, Rotter V.
Cold Spring Harb Perspect Biol. 2010 Feb;2(2):a001107.
Malkin D.
Genes Cancer. 2011 Apr;2(4):475-84.
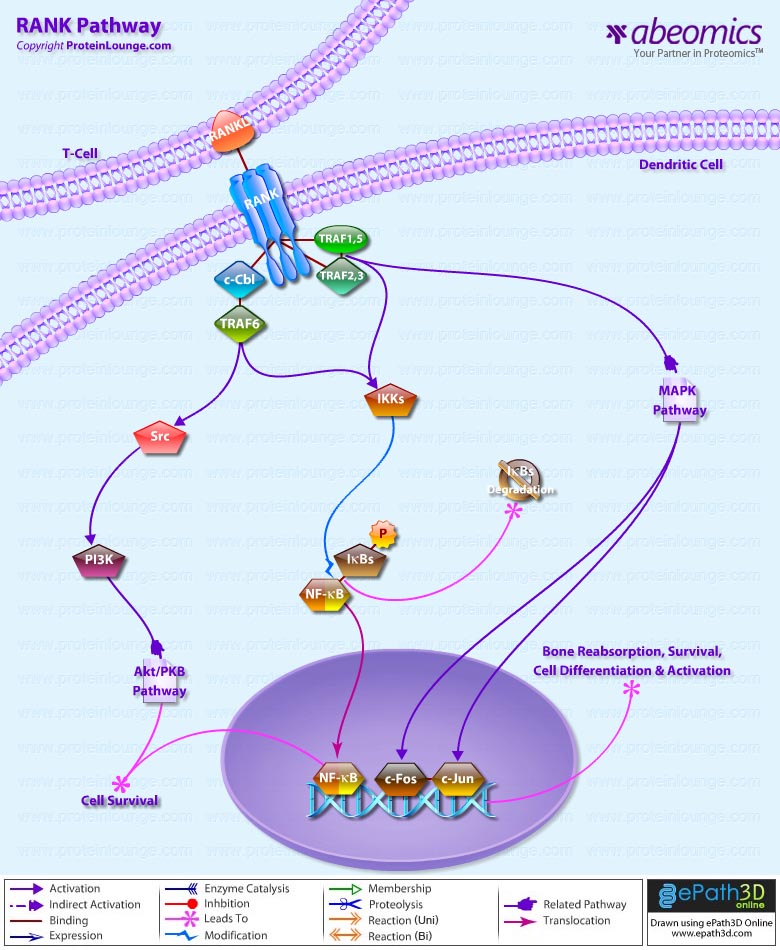
Bone remodeling and homeostasis is an essential function that regulates skeletal integrity throughout adult life in higher vertebrates and mammals. The structural and metabolic integrity of bone is maintained through the dynamic process of bone remodeling that results from the coordinate action of bone resorption by osteoclasts and the formation of new bone by osteoblasts. Regulation of bone remodeling occurs through multiple mechanisms that ultimately converge on the interaction of osteoclasts or their precursors with osteoblasts and bone marrow stromal cells. Key factors supplied by the stromal environment are cytokines such as IL-1, IL-11, IL-17, CSF1 (Colony Stimulating Factor-1) and the TNF receptor ligand family, RANKL (Receptor Activator of NF-kappaB Ligand; also known as OPGL, TRANCE, ODF and TNFSF11) and its receptor RANK (TNFRSF11A). Osteoclast activation is a critical cellular process for pathological bone resorption, such as erosions in Rheumatoid Arthritis or generalized bone loss (Ref.1 & 2).
RANKL is expressed on osteoblasts and bone marrow stromal cells, while its receptor RANK is expressed on osteoclast progenitors or mature osteoclasts. RANKL interacts with RANK via direct cell-cell contact, thereby promoting the differentiation, survival, and bone-resorbing capability of osteoclasts (Ref.3). Signaling through RANK involves the recruitment of cytosolic TRAFs (Tumor Necrosis Factor Receptor Associated Factors) TRAF1, TRAF2, TRAF3, TRAF5, and TRAF6, which in turn activate multiple signaling pathways. The association of RANK with TRAF2 induces activation of MAPK (Mitogen-Activated Protein Kinase), NF-KappaB and JNK (c-Jun N-Terminal Kinase), which leads to phosphorylation of c-Jun and activation of transcription factor, Activating Protein-1. Activation of NF-KappaB-inducing kinase, leads to activation of IKKs (I-KappaB kinases), which in turn phosphorylate serine residues on I-KappaBs (Inhibitor of Kappa Light Chain Gene Enhancer in B-Cells), targeting it for degradation in the proteasome. I-kappaB degradation exposes the NF-KappaB nuclear localization sequence, permitting its nuclear import. Within the nucleus, NF-KappaB acts in concert with other transcription factors to regulate gene expression (Ref.3). The RANK cytoplasmic tail associates with c-Src kinase, which activates the anti-apoptotic serine/threonine kinase Akt/PKB (Protein Kinase-B) through a signaling complex involving c-Src and TRAF6 (Ref.4). Moreover, RANK recruits c-Cbl family scaffolding proteins, and PI3K (Phosphatidylinositol-3 Kinase) in a ligand and Src-dependent manner. c-Src and TRAF6 interact with each other and with RANK following receptor engagement, and a deficiency in c-Src or the addition of Src family kinase inhibitors blocks RANK-mediated Akt/PKB activation in osteoclasts. TRAF6 enhances the kinase activity of c-Src leading to tyrosine phosphorylation of downstream signaling molecules such as c-Cbl, NF-KappaB and c-Fos, which is an important mediator of osteoclast differentiation (Ref.5).
RANKL and its receptor RANK are key regulators of bone remodeling, and are essential for the development and activation of osteoclasts. RANKL/RANK interactions regulate T-Cell/dendritic cell communications, dendritic cell survival, and lymph node formation. T-Cell–derived RANKL mediate autoimmune diseases, cancers, leukemia, asthma, chronic viral infections, and periodontal disease, which result in systemic and local bone loss and cartilage destruction in arthritis. RANKL and RANK are expressed in mammary gland epithelial cells, and they control the development of a lactating mammary gland during pregnancy. Inhibition of RANKL function via the natural decoy receptor OPG prevents bone loss in postmenopausal osteoporosis and cancer metastases and is useful to treat osteoporosis, crippling in arthritis, osteopenic disorders such as Paget’s disease, and bone loss and pain associated with bone metastases (Ref.6). This system provides an unexpected molecular paradigm that links bone morphogenesis, T-Cell activation, and the organization of lymphoid tissues, with mammary gland formation required for the survival of mammalian species. Modulation of these systems provides a unique opportunity to design novel therapeutics to inhibit bone loss in arthritis, periodontal disease, and osteoporosis (Ref.5).
References
1.Bone remodelling at a glance.
Crockett JC, Rogers MJ, Coxon FP, Hocking LJ, Helfrich MH.
J Cell Sci. 2011 Apr 1;124(Pt 7):991-8.
2.Osteoblast physiology in normal and pathological conditions.
Neve A, Corrado A, Cantatore FP.
Cell Tissue Res. 2011 Feb;343(2):289-302. Epub 2010 Dec 1.
Dougall WC.
Clin Cancer Res. 2012 Jan 15;18(2):326-35. Epub 2011 Oct 26.
Mellis DJ, Itzstein C, Helfrich MH, Crockett JC.
J Endocrinol. 2011 Nov;211(2):131-43. doi: 10.1530/JOE-11-0212. Epub 2011 Sep 8.
5.RANK(L) as a key target for controlling bone loss.
Leibbrandt A, Penninger JM.
Adv Exp Med Biol. 2009;647:130-45.
6.RANK/RANKL: regulators of immune responses and bone physiology.
Leibbrandt A, Penninger JM.
Ann N Y Acad Sci. 2008 Nov;1143:123-50.
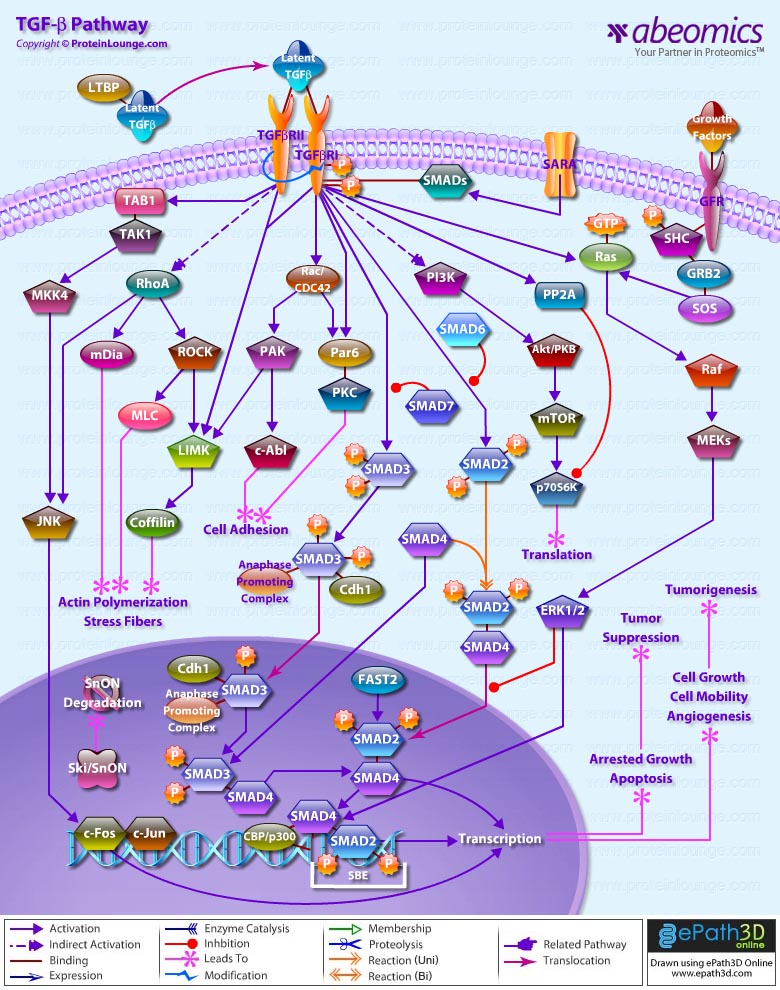
Cell proliferation in somatic tissues, specification of cell fate during embryogenesis, differentiation and cell death are controlled by a multitude of cell–cell signals and loss of this control has devastating consequences. Prominent among these regulatory signals is the TGF-Beta (Transforming Growth Factor) super family, which comprises a large and diverse group of polypeptide morphogens including the prototype of the family–the TGF-Beta themselves as well as the BMPs (Bone Morphogenetic Proteins), and the GDFs (Growth and Differentiation Factors) (Ref.1). The members of the TGF-Beta family are expressed in distinct temporal and tissue-specific patterns and therefore play an important role in the development, homeostasis and repair of most tissues in organisms. All immune cell lineages, including B-Cell, T-Cell and dendritic cells as well as macrophages secrete TGF-Beta, which negatively regulates their proliferation, differentiation and activation by other cytokines. Thus, TGF-Beta is a potent immunosuppressor and perturbation of TGF-Beta signaling is linked to autoimmunity, inflammation and cancer (Ref.2).
Before binding to its receptors, TGF-Beta is activated from a large latent complex comprised of LTBP and LAP (Latency Associated Peptide). Ligand binding to the Type II receptor (TGF-BetaRII) leads to recruitment of Type I receptor (TGF-BetaRI) in the highly conserved juxtamembrane region, known as the GS domain. The activated TGF-BetaRI then phosphorylates its downstream targets, the members of the SMAD (Sma and Mad Related Family) family of signal transducers, SMAD2 and SMAD3 (Ref.3). They form hetero-oligomeric complexes with SMAD4 and translocate to the nucleus, where they interact at the promoter with other transcription factors at DNA sequence-specific binding sites ATF2 (Activating Transcription Factor-2) and SBE (SMAD Binding Element) to regulate gene expression. The heteromeric SMAD complex also interacts with transcriptional coactivators and corepressors like p300 and CBP (CREB Binding Protein) to connect the SMAD-TF complex with the basal transcriptional machinery and mediate the biological effects of TGF-Beta (Ref.3). SMAD2-SMAD4 complexes regulate transcriptional responses by specifically interacting with DNA-binding proteins such as FAST2 (Forkhead Activin Signal Transducer-2). Some of the activated target genes stimulate tumorigenesis while others suppress tumorigenesis. SMAD2 is multiply ubiquitinated later and selectively targeted for proteasome-dependent degradation (Ref.5). Over expression of SMAD7 inhibits phosphorylation of SMAD2 and SMAD3 by activated TGF-BetaRI. SMAD6 is quite different in structure from the other SMAD proteins and forms stable associations with TGF-BetaRI. It interferes with the phosphorylation of SMAD2 and the subsequent heteromerization with SMAD4, but does not inhibit the activity of SMAD3. The specificity of receptor–SMAD interactions is dictated by discrete structural elements in the receptor kinase domain and the MAD homology domain of the SMAD. Prior to activation, receptor regulated SMADs are anchored to the cell membrane by factors like SARA (SMAD Anchor for Receptor Activation) that brings the SMADs into proximity of the TGF receptor kinases (Ref.4).
TGF-Beta also induces other non-SMAD signaling pathways, which include activation of several MKKs (MAP kinase Kinase) and MEKs (MAPK/ERK Kinase) pathways (JNK/SPAK, p38, and ERK1/2) through upstream mediators RhoA, Ras, TAK1 (TGF-Beta Activated Kinase), TAB1 (TAK1 Binding Protein) and the proteins XIAP (Xenopus Inhibitor of Apoptosis), HPK1 (Haematopoietic Progenitor Kinase-1) are also involved in this link (Ref.2). Because of its critical role in cell fate determination, TGF-Beta signaling is subject to many levels of positive and negative regulation, targeting both the receptors and the intracellular mediators. Among the negative regulators of SMAD function are two highly conserved members of the Ski family of proto-oncoproteins c-Ski and c-SnON that antagonizes TGF-Beta signaling through direct interactions with the SMAD2/SMAD3 and SMAD4 (Ref.6) and later degrade releasing SMADs to regulate transcription. SMAD3 recruits the APC (Anaphase-Promoting Complex) and Cdh1 (Cadherin-1) to SnON, thus providing an alternative mechanism to target SnON for ubiquitination and degradation (Ref.3). Besides the direct induction or repression of target gene expression, TGF-Beta also induces a variety of complex cellular responses, depending on the cell type, most notably growth arrest in late G1 involving PI3K (Phosphatidylinositol-3-Kinase) and PP2A (Protein Phosphatase-2A), changes in differentiation programs, and apoptosis (Ref.3). Other growth factors also regulate TGF-Beta mediated signaling through GRB2 (Growth Factor Receptor-Bound Protein-2)-SOS activation of Ras.
TGF-Beta are possibly the most pleiotropic secreted proteins functioning as morphogens mediating several physiological processes including hematopoiesis, regulation of hormone secretion, in immune response, angiogenesis, tissue morphogenesis and regeneration, and bone induction and modulation. With respect to bone induction, TGF-Beta induces substantial endochondral bone formation in a muscle tissue site but limited bone formation in a bony site (Ref.1). Failure or dysregulation of TGF-Beta signaling is involved in the development of several diseases, such as hematological malignancies, i.e., Leukemia, Haemorragic Telangiectasia, Chondrodysplasias, impaired wound healing, neurodegenerative conditions, developmental disorders and Pulmonary Hypertension. Genetic or epigenetic loss of TGF-Beta signaling promotes tumorigenesis via suppression of the immune system and changes in cell differentiation of epithelial tumor cells, a phenomenon termed EMT (Epithelial Mesenchymal Transdifferentiation) (Ref.2). In addition, recent work has implicated TGF-Beta in other processes involved in tumor inhibition including maintenance of genomic stability, induction of senescence, suppression of telomerase activity and prevention of inappropriate angiogenesis. More recently in certain patients infected with HIV1 (Human Immunodeficiency Virus Type-1), increased levels of TGF-Beta promoted the production of virus and also impaired the host immune system suggesting a possible role in HIV1 viral gene regulation and pathogenesis (Ref.7).
References
Ripamonti U, Ferretti C, Teare J, Blann L.
J Craniofac Surg. 2009 Sep;20(5):1544-55.
2.Regulation of TGF-beta signaling by Smad7.
Yan X, Liu Z, Chen Y.
Acta Biochim Biophys Sin (Shanghai). 2009 Apr;41(4):263-72.
3.Meeting report: signaling schemes for TGF-beta.
Roberts AB, Derynck R.
Sci STKE. 2001 Dec 18; 2001(113): PE43.
4.Tgf-Beta superfamily receptors-targets for antiangiogenic therapy?
Otten J, Bokemeyer C, Fiedler W.
J Oncol. 2010;2010:317068. Epub 2010 May 13.
5.Regulation of TGF-beta signaling by Smad7.
Yan X, Liu Z, Chen Y.
Acta Biochim Biophys Sin (Shanghai). 2009 Apr;41(4):263-72.
6.Ski and SnoN, potent negative regulators of TGF-beta signaling.
Deheuninck J, Luo K.
Cell Res. 2009 Jan;19(1):47-57.
Stettner MR, Nance JA, Wright CA, Kinoshita Y, Kim WK, Morgello S, Rappaport J, Khalili K, Gordon J, Johnson EM.
J Gen Virol. 2009 Aug;90(Pt 8):2005-14. Epub 2009 May 6.