Tips for Detecting
Phosphoproteins by Western Blot
Protein phosphorylation is very important for protein post-translational modification that controls many cellular processes including metabolism, transcriptional and translation regulation, degradation of proteins, cellular signaling and communication, proliferation, differentiation, and cell survival [1]. Approximately 35% of human proteins are phosphorylated. Phosphoproteins are low in abundance, and, therefore, are, sometimes, challenging to detect and characterize by Western blot and other methods. Western blot is the most common method for detecting and quantifying phosphorylated proteins. Western for phosphoprotein is not really different from a regular Western blot but following factors need pay attention to in order to obtain consistently results. Although all factors are important, protein sample preparation is possibly the most critical and deserve in-depth exploring.
Keep phosphoproteins intact. Once cells are lysed, protease and phosphatase are immediately released into the cell lysate. The turnover rate of phosphatases is very rapid. A phosphorylated protein can be dephosphorylated in a matter of milliseconds [2]. To prevent proteins from being degraded and dephosphorylated, the cell lysate must be kept on ice. Protease and phosphatase inhibitor cocktails must also be added to cell lysis buffer prior to use. Soon after the protein concentration of cell lysate is determined, the cell lysate should be mixed with SDS-PAGE loading buffer as soon as possible because the loading buffer can also inhibit phosphatase activity.
Avoid Using Milk for Blocking. Milk contains casein, an abundant phosphoprotein that can cause high background when milk is used to block the membrane. When doing your Western blot, you should use an alternative blocking solution such as bovine serum albumin, or other non-milk blocking agent such as WB Blocking Solution (Cat# WA-008, Invent Biotechnologies).
Optimize Experimental Conditions. Phosphorylation is a key response to cellular signaling and the phosphorylation status of a protein changes dynamically. Some proteins are phosphorylated only under certain conditions such as chemical or biological stimulation. You need to optimize your conditions to find out when your protein is phosphorylated. For example, you may need to stimulate the cells prior to harvesting. Try different stimulation conditions and do time courses to find when the highest level of phosphorylation is achieved [3].
Multiplex the Phosphorylated Proteins and Have Proper Controls. A simple way to look at total protein versus phosphorylated protein is to look at both proteins on the same blot using fluorescent antibodies, this is also known as multiplexing. Using the right combination of secondary antibodies, you can easily examine and determine the phosphorylation state of a protein. Always include a control in which you probe for the total protein, regardless of phosphorylation state. Many companies sell paired antibodies that recognize unmodified or phosphorylated forms of proteins.
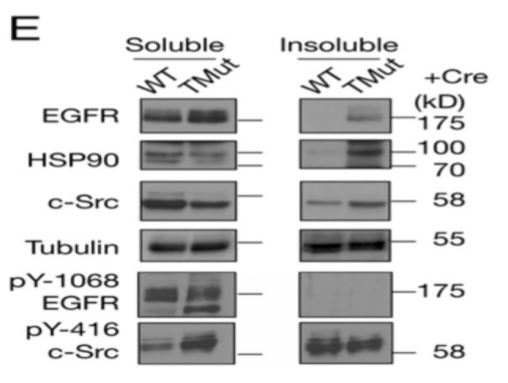
Sample Preparation. Sample preparation is a critical step for consistent detecting and quantifying phosphorylated proteins. Phosphoproteins may only accounts for a small fraction of total protein in a cell lysate depending upon cell growth condition and/or specific cell cycle stages. When a phosphorylated protein is not detected there are many possible reasons. However, few people question the methods used for sample preparation. The desired phosphorylated protein may or may not be present in the sample or present in the amount that is detectable by Western blot. The methods for protein extraction not only affect the extraction efficiency, it can also impact the quality of extracted proteins as discussed below. RIPA buffer is possibly one of the most commonly used cell lysis buffers for detection of phosphorylated protein by Western blot. However, it is known that the RIPA buffer generates two distinctive fractions in protein extraction e.g. RIPA soluble and RIPA insoluble fraction. Many proteins have been shown to be lost in the insoluble fraction [4,5]. Incomplete protein extraction may also affect the detection and quantification of phosphorylated proteins. Brady et al. [6] fractionated cell lysate into RIPA soluble and RIPA insoluble fractions and found that phosphorylated fragments of TDP-43 was detected in both fractions. Similarly, when mouse MEC cells were lysed by RIPA buffer, phosphorylated EGFR was detected in RIPA soluble fraction. However, phosphorylated c-Src was detected in both RIPA soluble and RIPA insoluble fractions. The signal in RIPA insoluble fraction is even stronger in the WT sample (Figure below) indicating that phosphorylated c-Sre was lost if only RIPA soluble fraction is used for the experiment [7]. If the phosphoprotein were quantified it would have been underestimated.
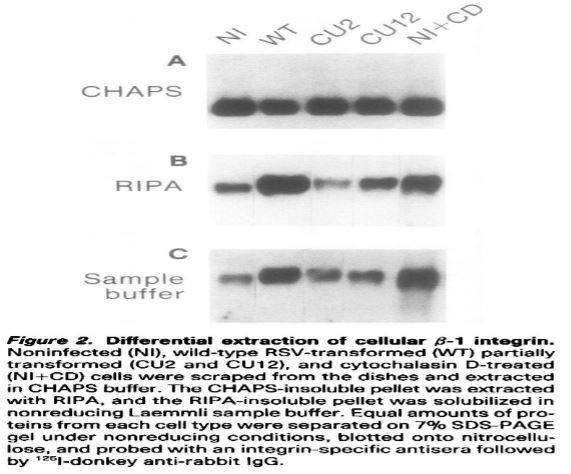
Beta integrin is a phosphoprotein, treatment with cytochalasin D causes an alter distribution of phosphorylated integrin between the Chaps soluble and insoluble fraction [8]). When a sequential protein extraction of primary chicken embryo fibroblasts was performed using Chaps, RIPA and Laemmli sample buffers, large amount of integrin was found in both Chaps insoluble and RIPA insoluble fractions for all five samples tested (Figure below).
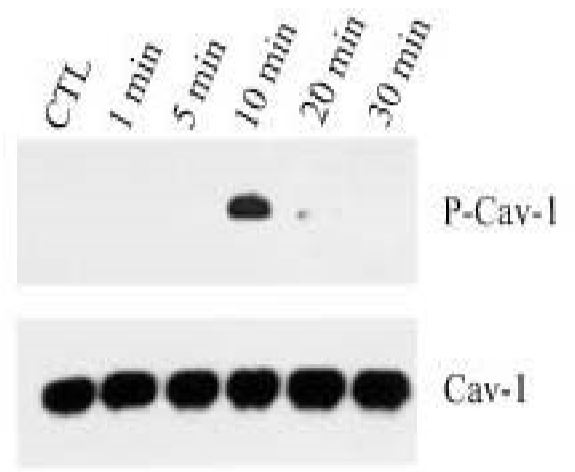
Tyrosine phosphorylation of caveolin-1 (P-Cav-1) was found to be mediated by activation of p38 mitogen activated kinase and c-Src kinase. Caveolin phosphorylation can be induced by 600 mM sucrose in 10 min. In this study [9], the cell lysate was prepared by Laemmli lysis buffer instead of RIPA buffer because this protein was found exclusively in RIPA insoluble fraction (see NIH3T3 cell lysate figure below).
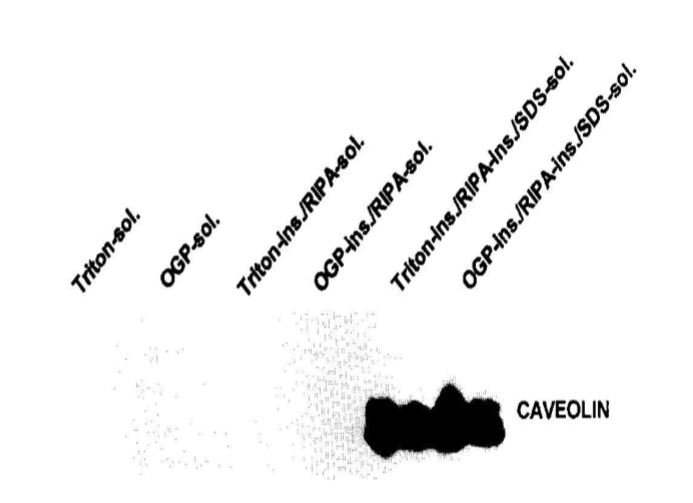
When NIH 3T3 cells were extracted by Triton-X-100 and RIPA buffer, caveolin is not detected in soluble fractions. It was exclusively found in insoluble fraction of these buffers [10 and figure below]. If RIPA soluble fraction were used in this case for detection of phosphorylation. The P-Cav-1 would have been missed.
For a given sample at a given time, the sum of phosphorylated and non-phosphorylated protein represents the total amount of this particular protein. An increase in amount of phosphorylated protein is usually accompanied by concomitant deduction in non-phosphorylated form of the protein as detected by Western blotting in the same blot. If a portion of protein is lost during protein extraction process, the ratio between phosphorylated and non-phosphorylated proteins may be altered resulting in biased data interpretation. Following data compared phosphorylation patterns of p-stat-3 and p-56 using two different protein extraction method (SD-001, a spin-column based protein extraction kit vs. RIPA buffer, un-published data. Courtesy of G. Szanda, NIH.NIAAA/LPS, USA).
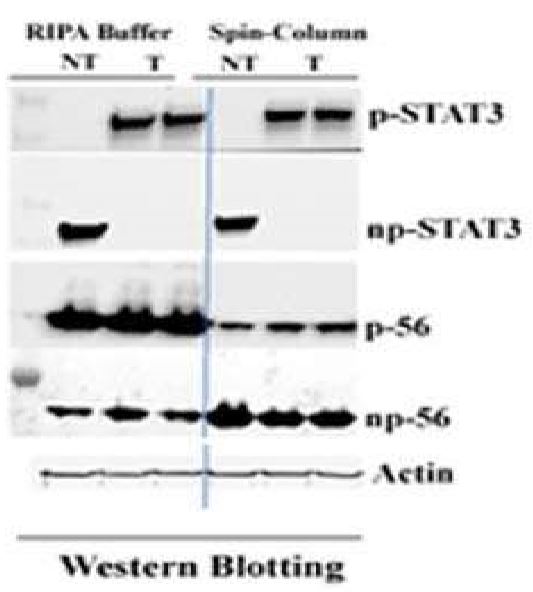
The results show that There is no difference between SD-001 and RIPA buffer for both treated (T) and Not treated (NT) samples in regard to the patterns of stat3. However, the phosphorylation pattern of p-56 is quite different. In SD-001 lysate, phosphorylated p-56 shows an increase from the base line level (NT) after a chemical stimulation (T) with a concomitant reduction in un-phosphorylated form, while RIPA buffer extracted sample is just the opposite. The phosphorylated p-56 in treated (T) sample is less than base line level (NT). This observation is against expected result. Although the precise reason is not clear, inadequate protein sample preparation could be a possible cause. In addition to protein loss during extraction process, RIPA buffer was also found to artificially increase the kinase activity and potentially affects protein phosphorylation [11]. As a summary, Detection and quantification of phosphorylated proteins by Western blotting is a common practice. Many factors can affect the results. When a phosphoprotein signal is not detected or weakly detected, improper protein extraction protocol may be responsible in addition to other potential problems discussed above. Therefore, selection of a proper sample preparation method is critical for successful detection and quantification of phosphoproteins.
References
1. Cohen P. (2000) The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem Sci. 25, 596-601.
2. Mansfield, C. et al. (2012) Stretch of contracting cardiac muscle abruptly deceases the rate of phosphate release at high and low calcium. JBC 287:25896-25705.
3. Tarrant, M. K. and Cole, P. A. (2009) The chemical biology of protein phosphorylation. Annu Rev Biochem. 78:797-825.
4. Li, Q. (2016) Pitfalls of protein extraction by RIPA buffer. Biotechniques. 61:327
5. Ngoka, L. CM. (2008) Sample prep for proteomics of breast cancer: proteomics and gene ontology reveal dramatic differences in protein solubilization preferences of radioimmunoprecipitation assay and urea lysis buffers. Proteome Science. 6:3
6. Brady, O. A. et al. (2011) Regulation of TDP-43 aggregation by phosphorylation and p62/SQSTM1. J. Neurochem. 116: 248-259.
7. Mukhopadhyay, C. et al. (2016) Casitas B-cell lymphoma (Cbl) proteins protect mammary epithelial cells from proteotoxincity of active c-Src accumulation. PNAS. 5: 8228-8237.
8. Haimovich, B. et al. (1991) Cellular partitioning of betta-1 integrins and their phosphorylated forms is altered after transformation by Rous sarcoma virus or treatment with cytochalasin D. Cell Regulation. 2:271-283.
9. Yan, S. R. et al. (1996) Activation of SRC family kinases in human neutrophils. Evidence that p58c-FGR and p53/56LYN redistribution to a triton X-100 insoluble cytoskeletal fraction, also enriched in the caveolar protein Caveolin, display an enhanced kinase activity. FEBS letter. 380:198-203.
10. Volonte, D. et al. (2001) Cellular stress induces the tyrosine phosphorylation of calveolin-1 (Tyr14) via activation of p38 mitogen-activated protein kinase and c-Src kinase. J. Biol. Chem. 276: 80948103.
11. DeSeau, V. et al. (Analysis of pp60c-src tyrosine kinase activity and phosphotyrosyl phosphatase activity in human colon carcinoma and normal human colon mucosal cells. J. Cellular Biochem. 35: 113-128.